Affinity Selection-Mass Spectrometry Analysis Service
Affinity selection-mass spectrometry (AS-MS) integrates protein affinity purification methods with small molecule mass spectrometry (MS) techniques, offering a non-biased and systematic approach for large-scale screening of small molecule ligands that interact with target proteins. This method has been successfully applied to the ligand screening of various soluble drug targets and is closely integrated with biochemical and cellular function experiments. It has tremendous potential in the discovery of novel lead compounds and also holds significant advantages in uncovering new targets within key signal transduction pathways. As high-resolution MS continues to improve in sensitivity and coverage, it is foreseeable that AS-MS will play an increasingly prominent role in fields such as the screening of natural active compounds targeting proteins and the regulation of protein functions by intracellular metabolites.
The AS-MS methods include affinity column MS (AC-MS), affinity capillary electrophoresis MS (ACE-MS), frontal affinity chromatography MS (FAC-MS), size exclusion chromatography (SEC) AS-MS, pulsed ultrafiltration (PUF) AS-MS, native MS screening (Naïve MS), also known as bio-affinity characterization MS (BACMS) and direct affinity selection MS, and magnetic microbead affinity selection screening (MagMASS). AS-MS adopts solution phase MS ionization technologies such as electric spray or atmospheric chemical ionization, which is compatible with online liquid chromatography (LC) and electrophoresis. Some AS-MS methods (e.g., SEC LC-MS and capillary electrophoresis MS (CE-MS) ) require online separation steps, while PUF AS-MS and MagMASS may opt for chromatographic separation.
The basic principle of AS-MS analysis involves obtaining the complex formed by the ligand and the target protein, separating this complex from unbound ligands in the solution using a purification method, and finally analyzing the ligands dissociated from the complex through LC-MS to identify their structures. Small-molecule ligands that specifically bind to target proteins are usually screened from a large pool of compounds, and such mixed compound screening approach can significantly increase screening throughput and reduce costs. In Affinity Selection-Mass Spectrometry (AS-MS), this most widely used methods currently include PUF AS-MS, SEC AS-MS, and MagMASS.
1. PUF AS-MS
The process of PUF AS-MS starts by incubating a combinatorial library mixture or extracts from plants, fungi, or microbial fermentation with a macromolecular receptor in solution. Typically, the concentrations of ligands and receptors are at the low micromolar level for screening, enabling the identification of ligands with dissociation constants in this range or better. If only higher affinity ligands need to be detected, the concentration of the receptor can be accordingly reduced. Upon achieving equilibrium, ultrafiltration is employed to separate the larger ligand-receptor complexes from the unbound compounds of lower mass. The selection of ultrafiltration membranes aims for the largest possible pore size while still retaining the macromolecular receptors, as larger pores facilitate faster ultrafiltration separation. The chemical composition of the ultrafiltration membranes is chosen to minimize nonspecific binding to the receptor and small molecules. The ligand-receptor complexes are then disrupted to release the bound ligands. This can be accomplished using organic solvents and/or changes in pH levels, such as using volatile organic acids (e.g., formic acid) with 90% methanol or acetonitrile. If the receptor is to be reused, conditions that might denature it should ideally be avoided.
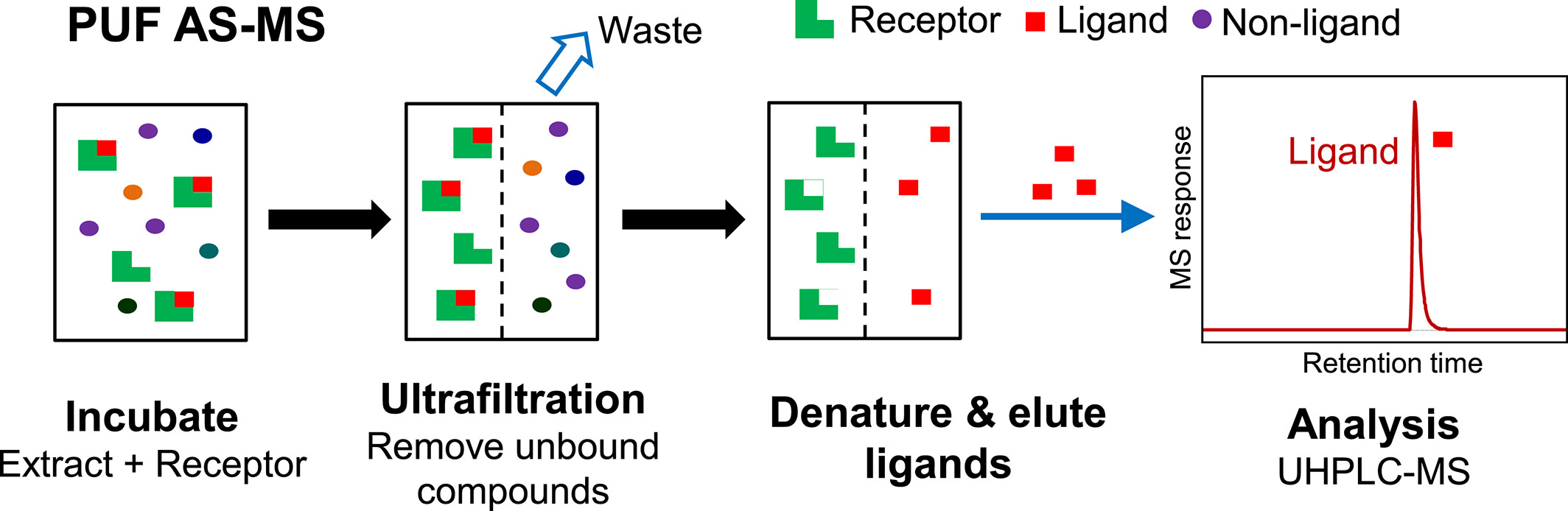
Figure 1. PUF AS-MS Process [3]
2. SEC AS-MS
SEC AS-MS is a solution-phase affinity screening method that utilizes SEC to separate ligand-receptor complexes from unbound compounds based on their size. Initially, the receptor is incubated with a mixture of potential ligands at concentrations similar to those used in PUF AS-MS. The ligand-receptor complexes are then separated using SEC, during which the large molecular complexes elute first, while the lower mass unbound compounds are retained within the small pores of the stationary phase.
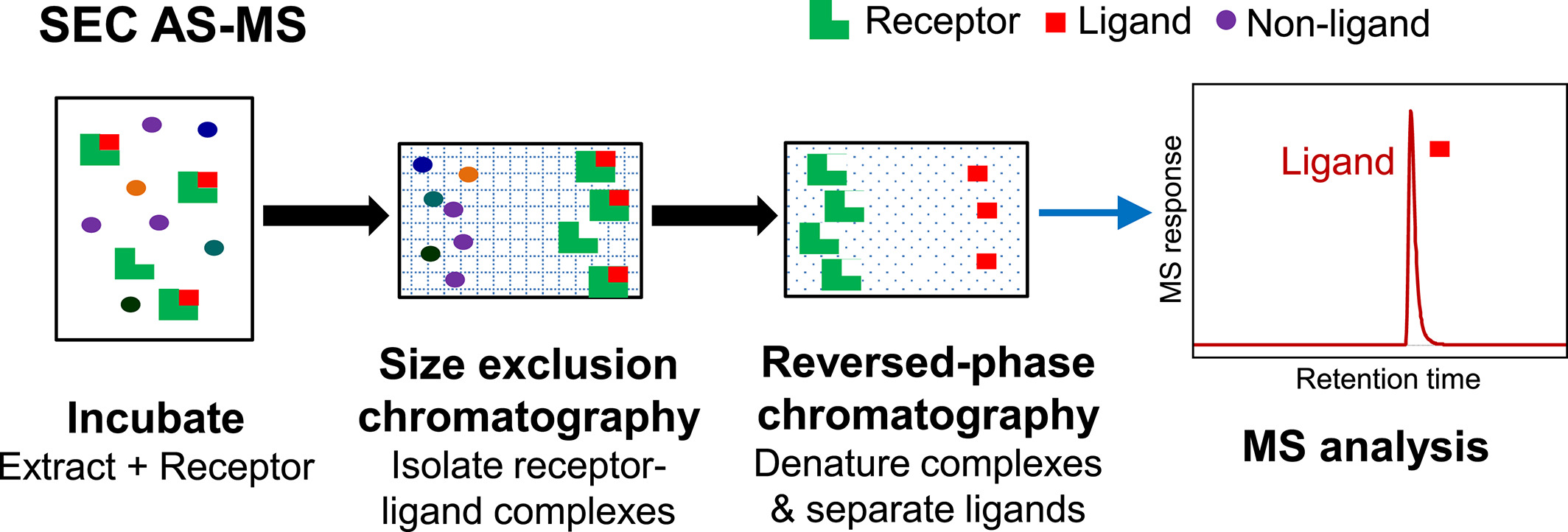
Figure 2. Basic Process of SEC AS-MS [3]
3. MagMASS
The MagMASS receptor is fixed on a solid support of magnetic microspheres, and depending on the chemical properties of the magnetic bead surface, the fixation of the receptor may involve covalent or non-covalent attachment. Functional groups on the surface of magnetic beads include primary amines, thiols, aldehydes, hydroxyls, and carboxylic acids. If the receptor possesses multiple reactive sites, it can be immobilized in various orientations, some of which may modify or sterically hinder the ligand-binding sites. Similar to the receptor of PUF AS-MS and SEC AS-MS, the receptor in MagMASS may include macromolecules such as proteins, RNA, or DNA. However, unlike other methods, low molecular weight molecules can also be immobilized to be researched such as fragment-based lead discovery. One advantage of MagMASS over solution-phase AS-MS methods is that it can stabilize the receptor.
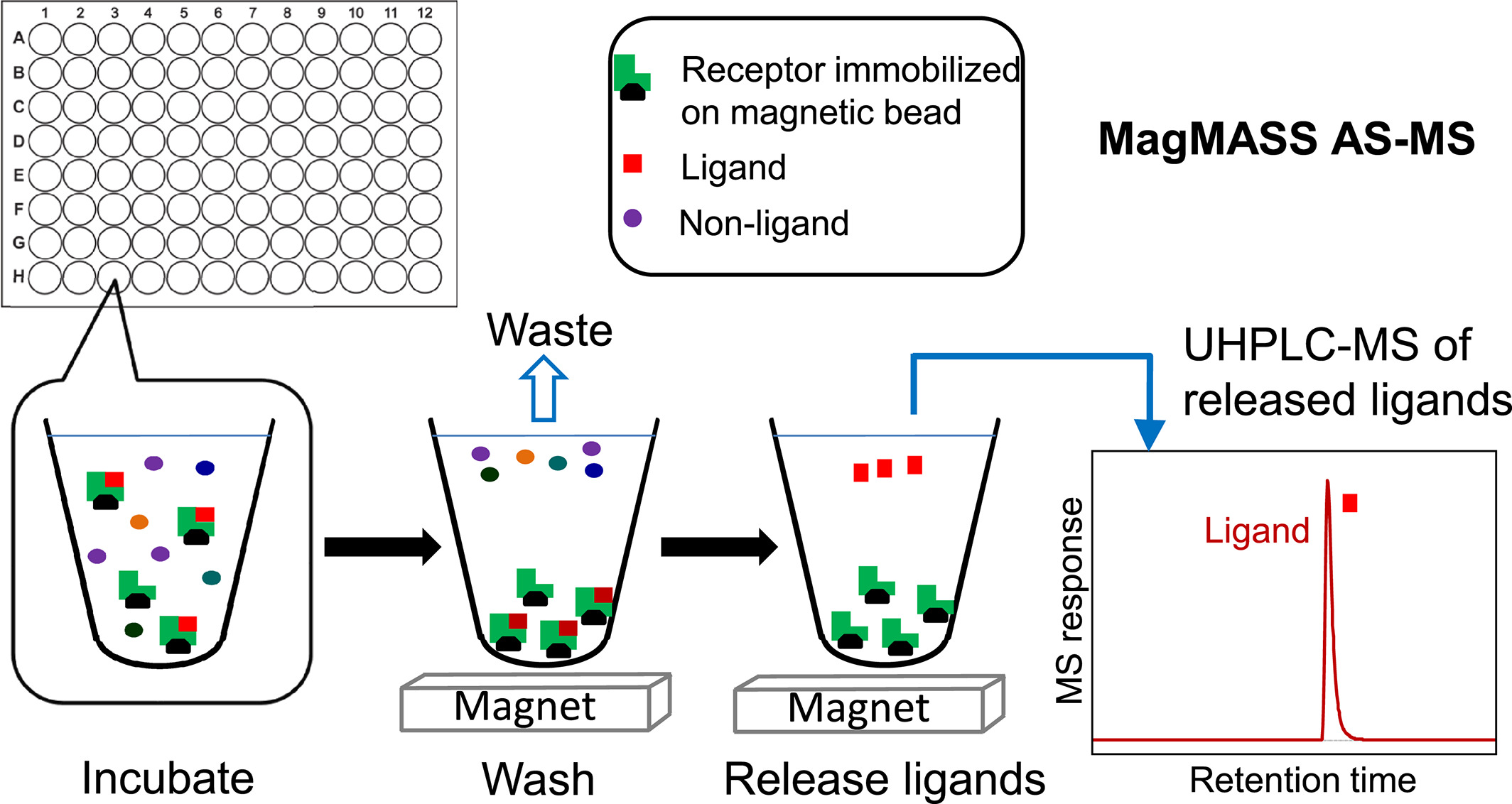
Figure 3. Basic Process of MagMASS AS-MS [3]
Analysis Workflow
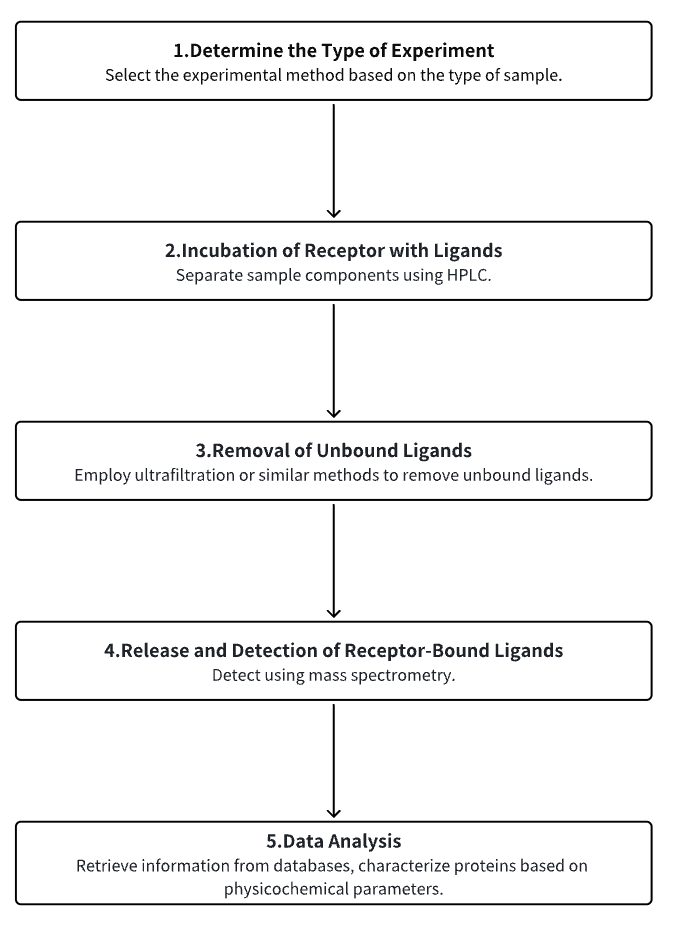
1. Determine the Type of Experiment
2. Incubation of Receptors with Ligands
3. Removal of Unbound Ligands
4. Release and Detection of Receptor-Bound Ligands
5. Data Analysis
Service Advantages
1. Detection of multiple sample types (protein & protein, protein & peptide, protein & small molecule).
2. A variety of experimental service offerings, including PUF AS-MS, SEC AS-MS, and MagMASS.
3. High-reliability detection results and comprehensive bioinformatics analysis.
Sample Results
1. The application of AS-MS in the discovery of anti-SARS-CoV-2 compounds. [4]
MS-based drug discovery methods utilizing affinity separation steps are suitable for screening complex mixtures of compounds, including natural product extracts and compounds in combinatorial libraries. These methods involve the binding of ligands to pharmacological receptors as affinity interactions, followed by the separation of ligand-receptor complexes from unbound inactive compounds in the mixture. The ligands extracted through affinity are then characterized and identified using MS.
MagMASS is applied to discover ligands for the SARS-CoV-2 spike protein. When the spike protein S1 subunit is immobilized on magnetic beads, the immobilized receptor is incubated with plant extracts containing potential ligands to reach an equilibrium condition. Magnets are used to capture the magnetic beads with ligand-receptor complexes, which are then washed with a binding buffer to remove unbound compounds. Organic solvents are used to denature the S1 protein and release the ligands for analysis by Ultra Performance Liquid Chromatography-MS (UHPLC-MS).
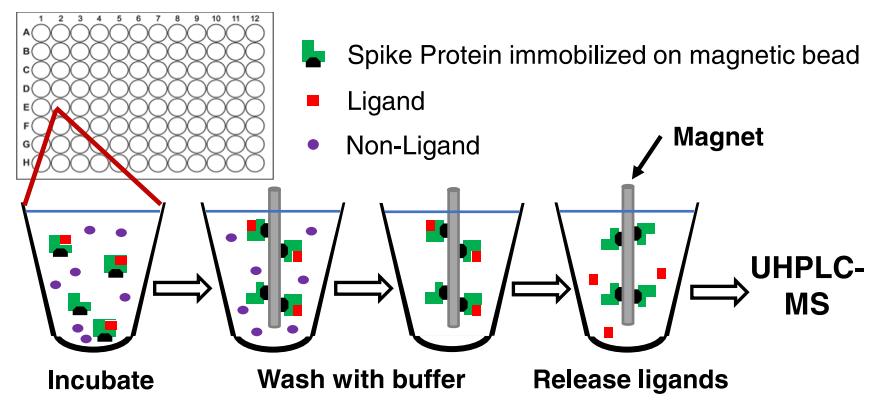
Figure 4. MagMASS for Ligands of the SARS-CoV-2 Spike Protein
Naïve MS screening is applied to discover SARS-CoV-2 replication inhibitors. After incubating the receptor with a mixture of potential ligands in a low molar concentration volatile aqueous buffer (such as 3 mM ammonium acetate), the desorbed receptor-ligand complexes are measured using electrospray ionization MS (ESI-MS) on an ultra-high-resolution MS. During the ESI-MS analysis, high mass receptor ions are detected as a series of multiply charged species (black), while the ligand-receptor complexes appear as a series of slightly higher m/z value multiply charged peaks, corresponding to the receptor plus the attached ligand (red).
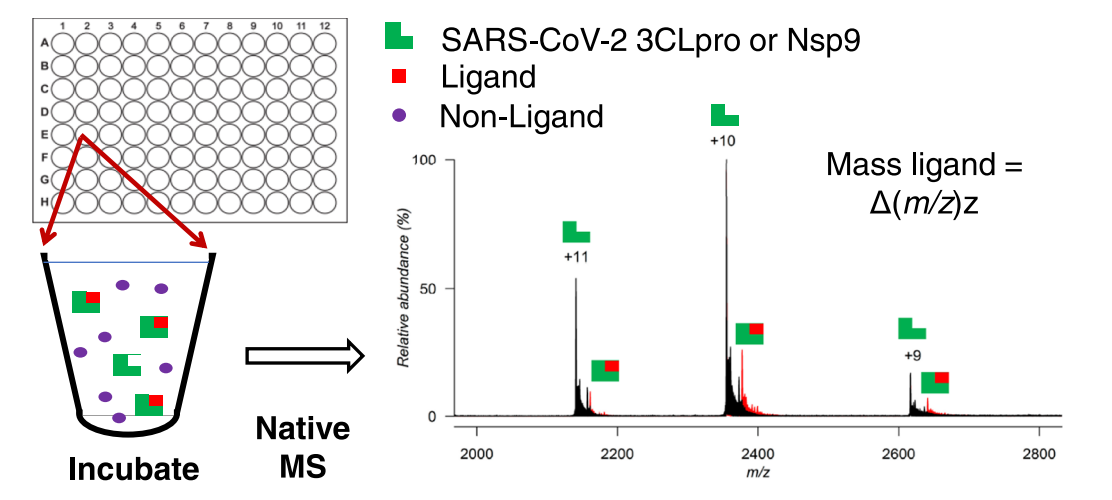
Figure 5. MagMASS for Ligands of the SARS-CoV-2 Spike Protein
2. Mitochondria-based centrifugal ultrafiltration (CU) LC-MS method for screening mitochondria-targeted bioactive components from complex matrices. [5]
Mitochondria represent critical pharmacological targets within cells, given their pivotal role in orchestrating complex biological processes including energy generation, apoptosis, and lipid metabolism. Damage to these organelles is implicated in a myriad of human pathologies. To expedite the search for naturally occurring bioactive compounds targeting mitochondria, researchers have developed an efficient screening method that combines CU with LC-MS, termed the screening method for mitochondria-targeting bioactive constituents (SM-MBC). Besides, this approach is compatible with the search for mitochondrial-targeting compounds from complex matrices such as herbal extracts. Using an optimized mitochondrial isolation scheme, functionally active, structurally intact, and pure mitochondria were obtained from rat myocardial tissue. Following the evaluation of the method's reliability with thiabendazole (TZ), rotenone (RN), amiodarone (AR), and trimeprazine (TD) as positive controls, this method was successfully applied to extract bioactive components from Polygoni Cuspidati Rhizoma et Radix (PCRR) and Scutellariae Radix (SR). LC-MS analysis identified 19 bioactive compounds, of which 17 were novel mitochondrial-targeting compounds. The activity of 9 out of these 19 compounds was confirmed through in vitro pharmacological assays.
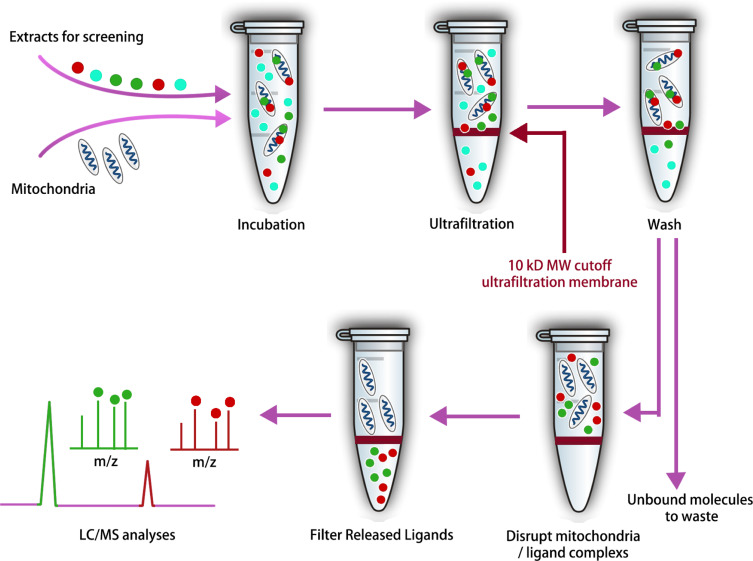
Figure 6. Ultrafiltration MS Workflow for the Identification of Mitochondrial-Targeted Bioactive Components From Complex Mixtures
3. AS-MS identifies the optimal binding ligands for the retinoid X receptor (RXR). [6]
Nuclear receptors, such as the RXR, are proteins that regulate countless cellular processes. Molecules acting as RXR agonists are of particular interest for the prevention and control of carcinogenesis. Most of these ligands have an acidic moiety, considered key for RXR activation. This study presents the design, synthesis, and biological evaluation of acidic and non-acidic indenoisoquinolines as novel RXR ligands, identifying critical features of indenoisoquinoline toxicants. The affinity ranking of synthesized indenoisoquinoline toxicants, among others in the library for RXR-α, was correlated with activity in cell-based RXR transcription induction assays, using PUF AS-MS.
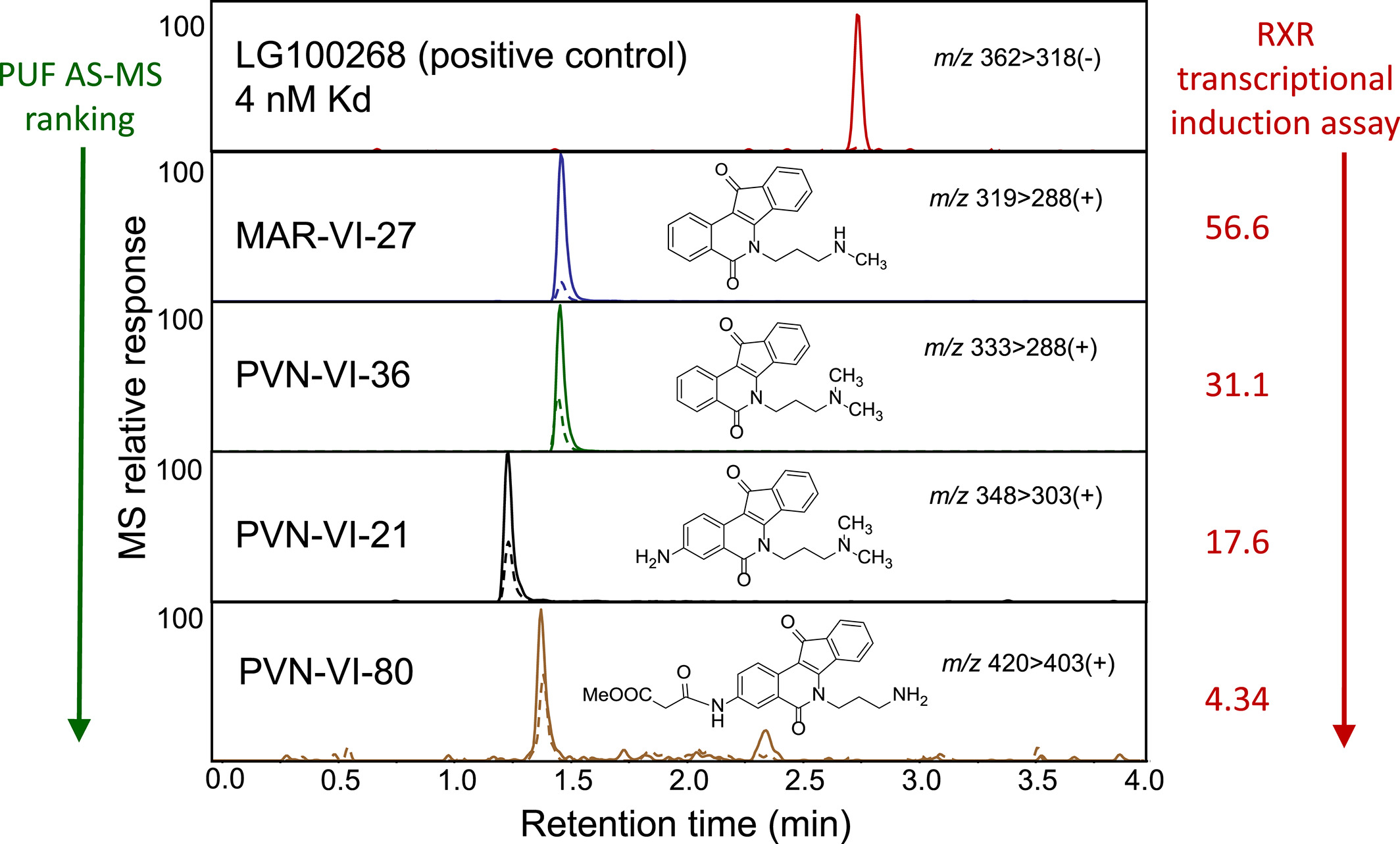
Figure 7. Ranking Ligands by Binding Affinity
Sample Submission Requirements
1. Provide pre-purified samples whenever possible.
2. Try to minimize impurity contamination.
Services at MtoZ Biolabs
1. Complete Experimental Protocol
2. Relevant Instrument Settings
3. Raw MS Data
4. Detailed Report on Compounds Bound to the Receptor. (including identification list, affinity ranking, etc.)
Applications
1. Natural Drug Screening
Currently, most natural product researchers employ bioassay-guided isolation processes, which is extremely time-consuming. Typically, pharmacological activity assays are conducted on plants, fungus, or microbial extracts or fractions. If an extract shows a positive response, chromatographic methods are used to fractionate it. The activity of individual fractions is assessed, and the process of fractionation and activity determination is repeated. This iterative process continues until the active compounds are isolated for spectroscopic identification. In contrast, AS-MS offers a faster alternative for screening complex natural product extracts.
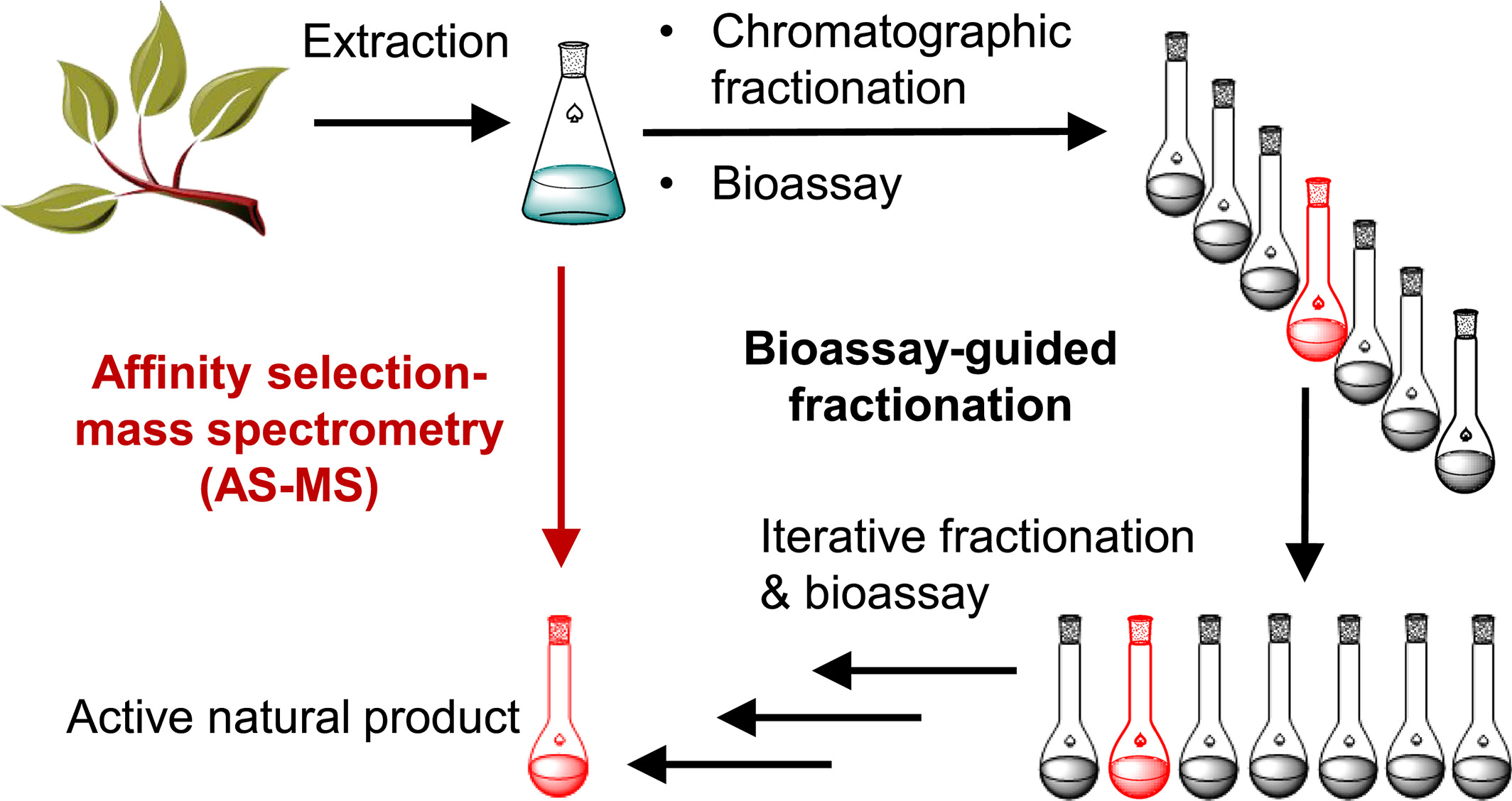
Figure 8. AS-MS Enables Rapid Screening of Active Components in Natural Products [7]
2. Small Molecule Drug Discovery
Identifying hit compounds is the first step in small molecule drug discovery, and most conventional high-throughput screening (HTS) assays, such as enzymatic or G protein-coupled receptor (GPCR) signaling assays, are based on the functional activity of targets. However, the number of drug targets that can be developed using functional HTS is limited, and many promising drug targets cannot be developed into HTS assays. Affinity-based screening techniques have been developed to address this situation. Compared to traditional biochemical HTS technologies, AS-MS is one of the most commonly used methods applicable to any drug target.
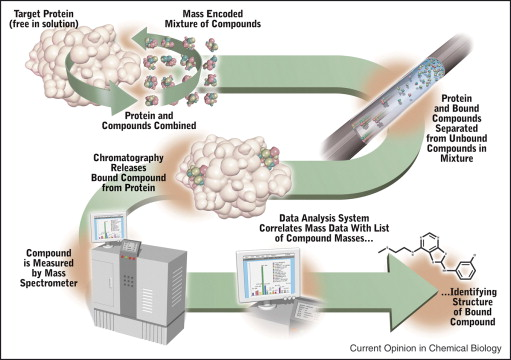
Figure 9. Schematic Diagram of AS-MS Screening for Small Molecule Drugs [8]
FAQ
Q1: There are many types of AS-MS, and what are the characteristics of each?
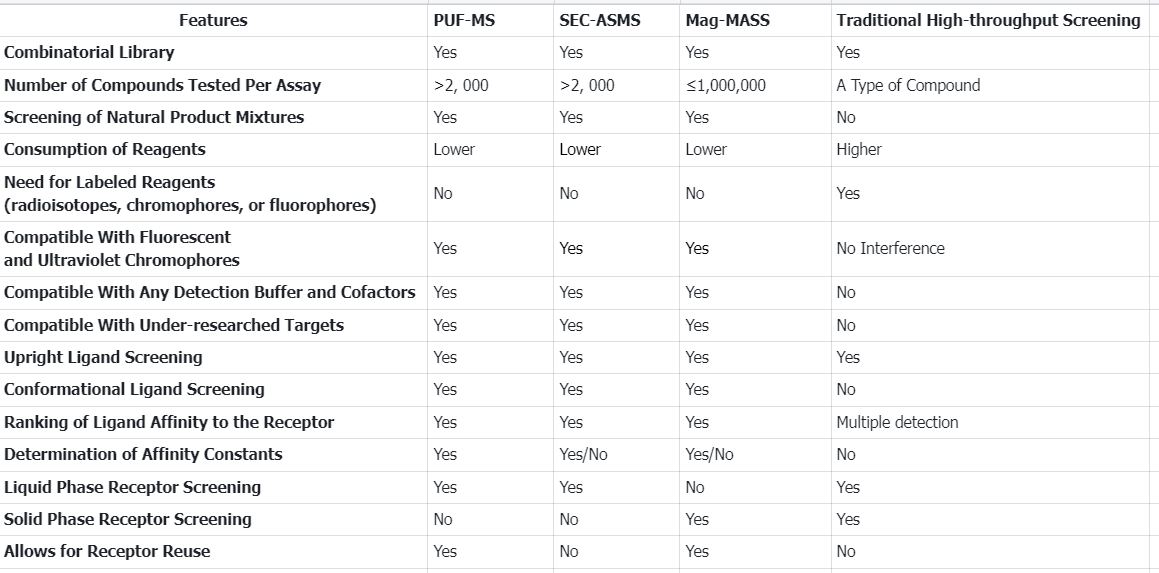
References
[1] Chan HCS, Shan H, Dahoun T, Vogel H, Yuan S. Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol Sci. 2019 Aug;40(8):592-604. doi: 10.1016/j.tips.2019.06.004. Epub 2019 Jul 15. Erratum in: Trends Pharmacol Sci. 2019 Oct;40(10):801. PMID: 31320117.
[2] Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. 2019 Sep 4;20(18):4331. doi: 10.3390/ijms20184331. PMID: 31487867; PMCID: PMC6769923.
[3] Prudent R, Annis DA, Dandliker PJ, Ortholand JY, Roche D. Exploring new targets and chemical space with affinity selection-mass spectrometry. Nat Rev Chem. 2021 Jan;5(1):62-71. doi: 10.1038/s41570-020-00229-2. Epub 2020 Oct 21. PMID: 37118102.
[4] van Breemen RB, Muchiri RN. Affinity selection-mass spectrometry in the discovery of anti-SARS-CoV-2 compounds. Mass Spectrom Rev. 2022 Aug 5:e21800. doi: 10.1002/mas.21800. Epub ahead of print. PMID: 35929396; PMCID: PMC9538385.
[5] Yang XX, Xu F, Wang D, Yang ZW, Tan HR, Shang MY, Wang X, Cai SQ. Development of a mitochondria-based centrifugal ultrafiltration/liquid chromatography/mass spectrometry method for screening mitochondria-targeted bioactive constituents from complex matrixes: Herbal medicines as a case study. J Chromatogr A. 2015 Sep 25;1413:33-46. doi: 10.1016/j.chroma.2015.08.014. Epub 2015 Aug 12. PMID: 26306914.
[6] Conda-Sheridan M, Park EJ, Beck DE, Reddy PV, Nguyen TX, Hu B, Chen L, White JJ, van Breemen RB, Pezzuto JM, Cushman M. Design, synthesis, and biological evaluation of indenoisoquinoline rexinoids with chemopreventive potential. J Med Chem. 2013 Mar 28;56(6):2581-605. doi: 10.1021/jm400026k. Epub 2013 Mar 8. PMID: 23472886; PMCID: PMC3623729.
[7] Muchiri RN, van Breemen RB. Affinity selection-mass spectrometry for the discovery of pharmacologically active compounds from combinatorial libraries and natural products. J Mass Spectrom. 2021;56(5):e4647. doi: 10.1002/jms.4647. Epub 2020 Sep 21. PMID: 32955158.
[8] Annis DA, Nickbarg E, Yang X, Ziebell MR, Whitehurst CE. Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr Opin Chem Biol. 2007 Oct;11(5):518-26. doi: 10.1016/j.cbpa.2007.07.011. Epub 2007 Oct 10. PMID: 17931956.
How to order?







