Proteomics Analysis Service
Proteomics analysis involves a comprehensive study of all proteins in an organism, tissue, or cell, focusing on their structure, function, expression levels, and interactions, to reveal their dynamic changes and molecular mechanisms in biological processes. Proteomics analysis encompasses protein identification, quantitative analysis, detection of post-translational modifications (PTMs), protein interaction network analysis, and functional pathway enrichment analysis, aiming to provide a thorough understanding of protein roles under various physiological and pathological conditions.
Proteomics analysis can uncover the connection between genes and phenotypes, elucidate the key molecular mechanisms underlying disease occurrence, progression, and treatment, and identify potential biomarkers and therapeutic targets. Through high-resolution mass spectrometry technologies and advanced bioinformatics tools, proteomics analysis addresses scientific challenges such as disease molecular mechanism analysis, drug target identification, and biomarker discovery, offering robust support for precision medicine, drug development, and fundamental life sciences research.
1. Protein Quantitation
Quantitative proteomics is divided into label-based methods and label-free methods. Labeled quantitative proteomics involves introducing light and heavy isotopes or chemical tags into samples to compare protein abundance under different conditions. Metabolic labeling (e.g., SILAC) uses labeled amino acids introduced during cell culture to metabolically label proteins, while chemical labeling (e.g., ICAT) modifies specific residues with specialized chemical tags. Enzymatic labeling incorporates 18O isotopes during the digestion process using heavy water. After labeling, samples are mixed in fixed ratios and analyzed by LC-MS/MS to quantify relative protein abundance based on differences in signal intensities of mass peaks. Label-based methods offer high precision and reproducibility, making them ideal for quantitative studies in complex samples. Label-free quantitative proteomics does not require additional labeling of samples and directly quantifies relative protein abundance by analyzing differences in peptide signal intensities or spectral counts using mass spectrometry. After separate digestion and mass spectrometry analysis of each sample, the relative abundance of proteins is calculated by comparing the signal intensities of the same peptides across different conditions. The label-free method has a simpler workflow and is suitable for large-scale samples or experiments where labeling is impractical, but it relies heavily on instrument stability and advanced data processing capabilities.
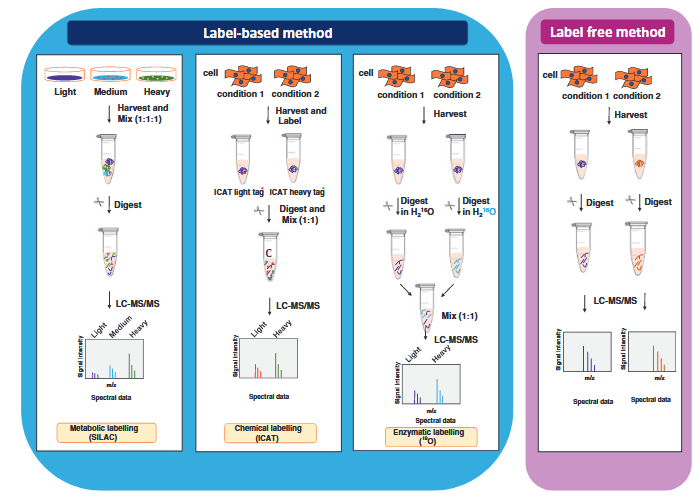
Anand, S. et al. Methods Mol Biol. 2017.
Figure 1. The Workflow of Quantitative Proteomics
2. Protein Identification
Protein identification holds a central position in proteomics analysis, serving as the foundation for understanding the composition, structure, and function of proteins within biological systems. The protein identification workflow consists of several key steps. First, proteins are extracted from biological samples such as animals, tissues, biofluids, or cells. Next, protein separation is performed to ensure effective differentiation of various proteins. Following this, proteolytic digestion using enzymes like trypsin breaks down proteins into peptides. Subsequently, chemical labeling (an optional step) can be applied to enhance the accuracy of quantitative analysis. The resulting peptides are then analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS), generating mass spectra. These spectra are used for peptide identification and quantification, which is further mapped to protein identification and quantification. Leveraging high-precision mass spectrometry technologies and advanced bioinformatics tools, protein identification reveals changes in protein expression, post-translational modifications, and protein-protein interactions under specific conditions, providing critical data for disease mechanism studies, biomarker discovery, and drug target screening.
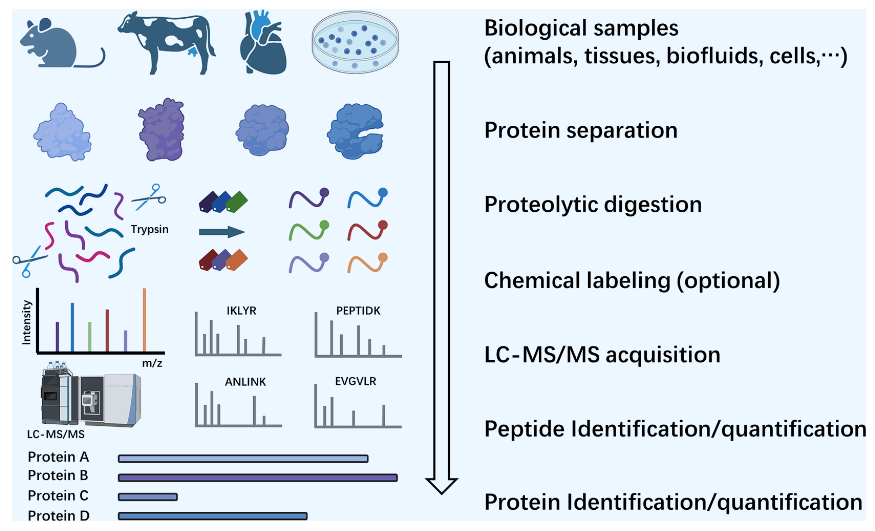
Wang, Z. et al. ACS Meas Sci Au. 2024.
Figure 2. Proteomics Strategies for Protein Identification
3. Protein Post-translational Modification Analysis
Based on this diagram, the post-translational modification (PTM) analysis workflow primarily follows three strategies: Bottom-up proteomics, Middle-down proteomics, and Top-down proteomics. In Bottom-up proteomics, protein samples undergo high-grade digestion to generate small peptides (8–30 amino acids). These peptides are separated and selected using LC-MS, followed by MS2 analysis to identify PTM sites. In Middle-down proteomics, proteins are subjected to medium-grade digestion to produce larger peptides (>30 amino acids). These peptides are separated via LC-MS, selected, and analyzed using MS2, preserving more sequence and modification information. In Top-down proteomics, proteins remain undigested and are analyzed as intact entities. They undergo protein separation, proteoform selection, and subsequent analysis via MS1 and MS2, allowing the identification of different proteoforms and PTM patterns. During PTM identification, the measured peptide mass is compared with the theoretically calculated sequence mass. Through the mass shift (Δm), specific modifications (e.g., acetylation, Δm = 42.01 Da) are identified and validated using PTM databases. This comprehensive workflow accurately identifies PTM sites, types, and their biological roles, revealing regulatory mechanisms in cellular processes.
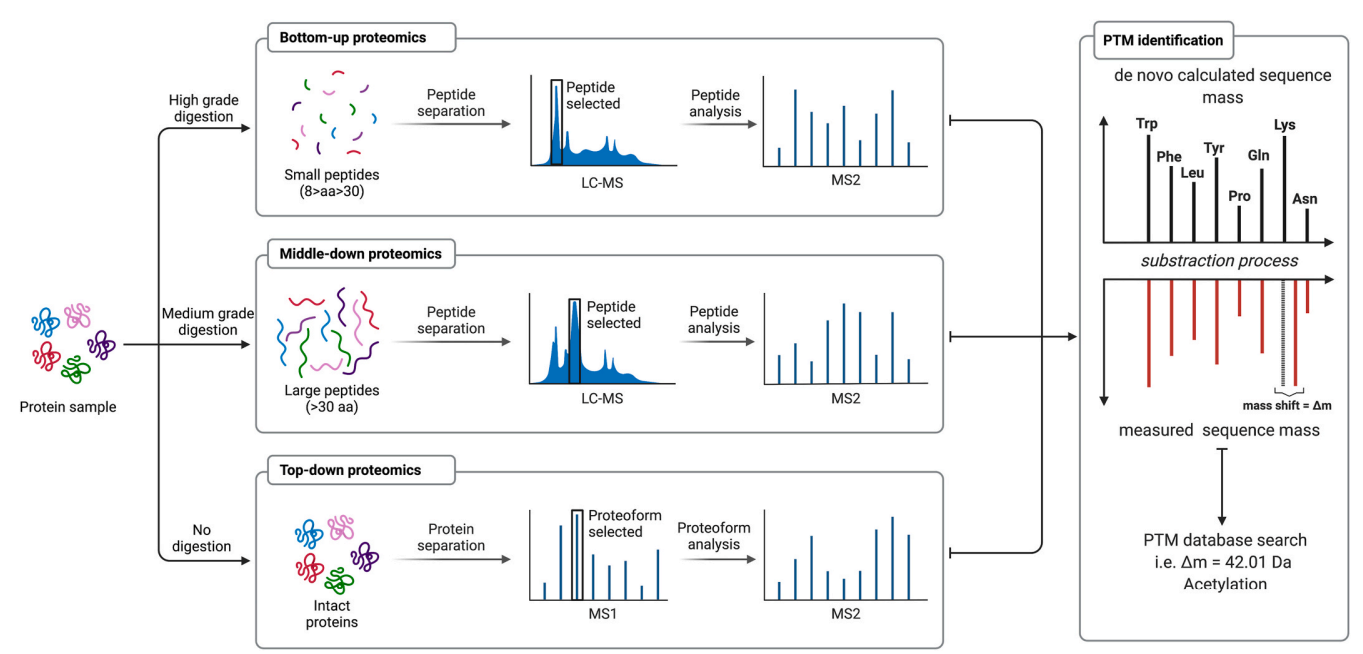
Hermann, J. et al. Mol Aspects Med. 2022.
Figure 3. The Workflow of Protein Post-translational Modification Analysis
4. Protein-Proten Interaction Analysis
Protein interaction analysis is a crucial approach for uncovering protein functions and regulatory networks within cells. The following are three main strategies:
(a) Affinity Purification: Using antibodies or tags (e.g., Strep-tag, TAP tags), target proteins (Bait) are enriched from cell lysates along with their direct or indirect interacting partners. These complexes are subsequently identified through mass spectrometry. This method is particularly suitable for analyzing stable protein complexes.
(b) Proximity Labeling: Techniques such as BioID or APEX are employed to covalently label neighboring proteins within a defined radius around the target protein using biotin. These biotin-labeled proteins are then purified via streptavidin and identified using mass spectrometry. This approach is ideal for capturing weak or transient protein interactions.
(c) Global Interactome Analysis: Proteins are analyzed globally through fractionation techniques (e.g., SEC, IEX). These methods enable the identification and quantification of protein complexes on a large scale. Finally, mass spectrometry and bioinformatics are used to integrate the data and construct a comprehensive protein interaction network (Interactome), revealing dynamic protein interactions in complex biological processes.
5. Sample Proteomics Analysis
Sample proteomics analysis involves a comprehensive investigation of the composition, structure, and function of proteins within a sample. First, protein extraction is performed to isolate total proteins from biological samples, such as cells, tissues, or biofluids, ensuring sample integrity and high quality. Next, protein digestion is carried out, typically using trypsin to break proteins into specific peptides, facilitating subsequent analysis. This is followed by MS/MS analysis, where liquid chromatography-tandem mass spectrometry (LC-MS/MS) is used to separate, identify, and quantify the peptides. Subsequently, data analysis is conducted using specialized bioinformatics tools to interpret the mass spectrometry data, identifying and quantifying proteins. Finally, a report generation step compiles the experimental results into a visualized report, revealing patterns of protein expression, interactions, and functional changes in biological processes. Sample proteomics analysis provides robust technical support for disease mechanism research, biomarker discovery, and drug target identification.
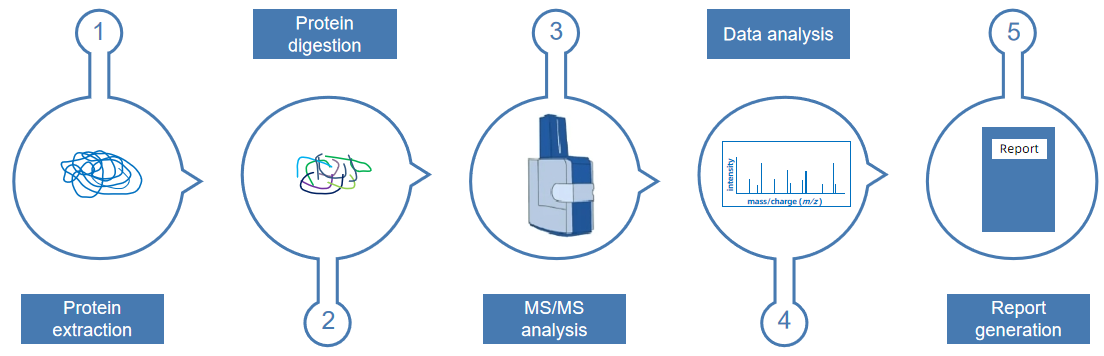
Figure 4. The Workflow of Sample Proteomics
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs leverages an advanced mass spectrometry platform, including the Thermo Fisher Orbitrap Fusion Lumos and Q Exactive HF mass spectrometers, combined with the Nano-LC nanoliter liquid chromatography system, to deliver high-precision and high-throughput proteomics analysis service that ensures comprehensive protein analysis and reliable data reproducibility for diverse research needs. We employ DDA (Data-Dependent Acquisition) and DIA (Data-Independent Acquisition) data acquisition modes, along with PRM (Parallel Reaction Monitoring) and MRM (Multiple Reaction Monitoring) detection modes, ensuring high sensitivity and specificity in target protein detection, as well as data accuracy and reproducibility. Our team of proteomics experts brings extensive project experience and the ability to efficiently address complex scientific challenges, providing clients with reliable, traceable experimental data and precise scientific reports to support the successful advancement of research projects. Below are some contents of our proteomics analysis service:
1. Protein Quatitation Service
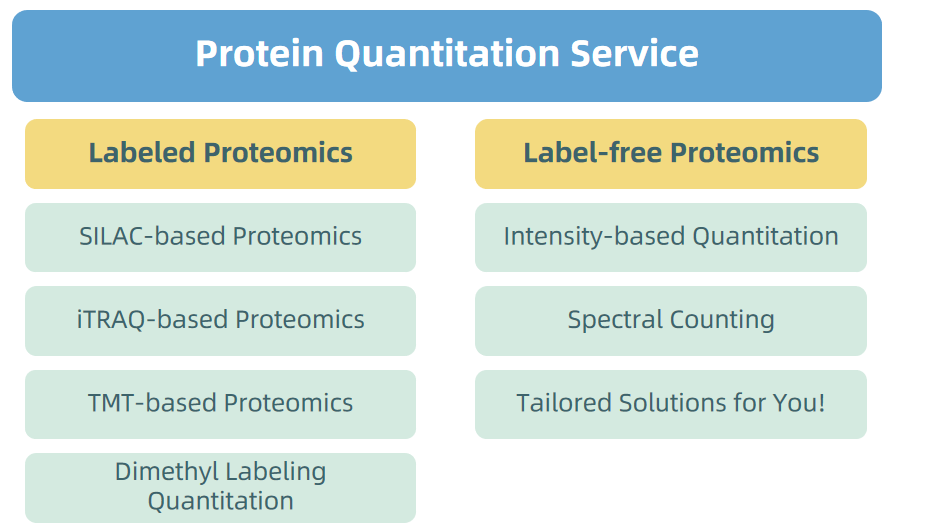
Figure 5. Protein Quantitation Service Provided by MtoZ Biolabs
2. Protein Identification Service
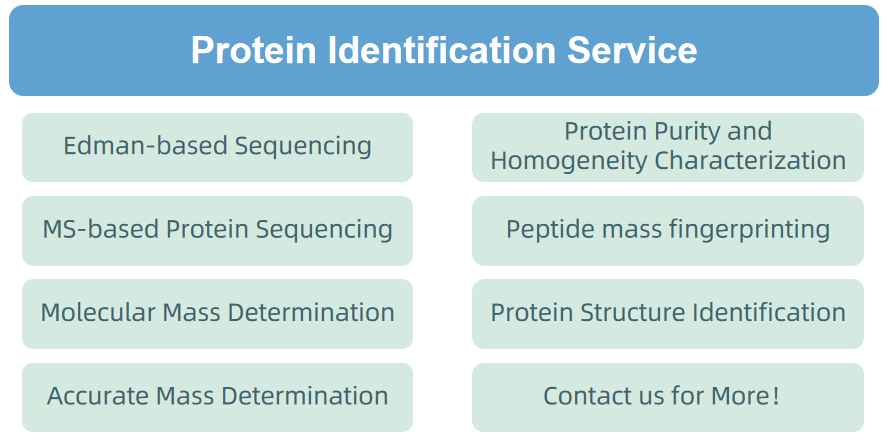
Figure 6. Protein Identification Service Provided by MtoZ Biolabs
3. Protein Post-translational Modification Analysis Service
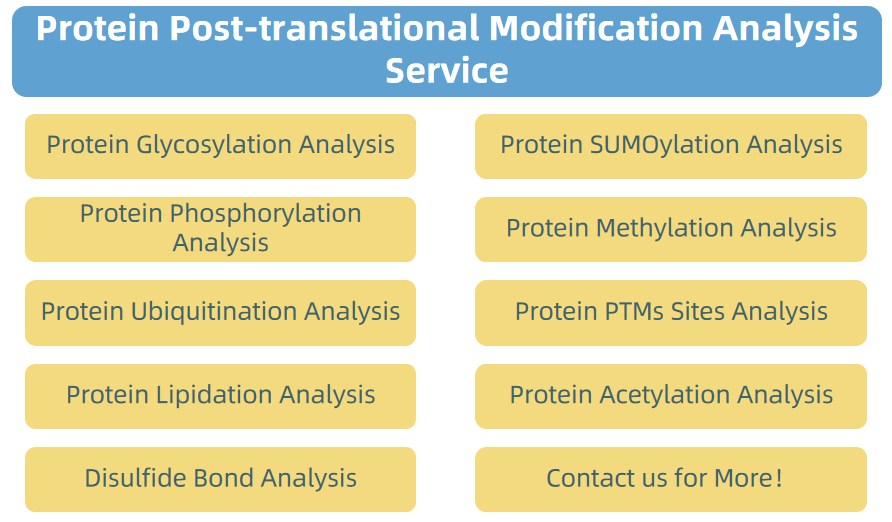
Figure 7. Protein Post-translational Modification Analysis Service Provided by MtoZ Biolabs
4. Protein-Proten Interaction Analysis Service
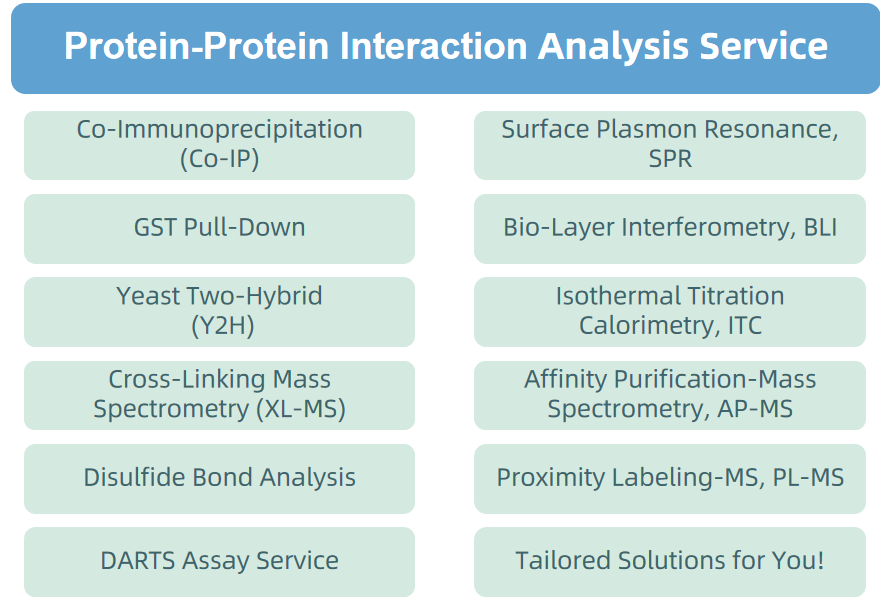
Figure 8. Protein-Proten Interaction Analysis Service Provided by MtoZ Biolabs
5. Sample Proteomics Analysis Service
Figure 9. Sample Proteomics Analysis Service Provided by MtoZ Biolabs
Service Advantages
1. Comprehensive Proteomics Data Acquisition Modes
Our proteomics analysis services combines Data-Dependent Acquisition (DDA) and Data-Independent Acquisition (DIA) modes to ensure high-throughput and extensive protein identification across samples of varying complexity. This approach minimizes data loss, enhances data integrity, and ensures reliable and comprehensive results.
2. High-Sensitivity and High-Specificity Targeted Protein Detection
Leveraging advanced Parallel Reaction Monitoring (PRM) and Multiple Reaction Monitoring (MRM) technologies, MtoZ Biolabs’ proteomics analysis services enable precise quantification of low-abundance proteins. These technologies offer highly specific detection of target proteins, ensuring both accuracy and reproducibility of the results.
3. Advanced Data Analysis and Bioinformatics Interpretation
Our proteomics analysis service allows for in-depth data mining, pathway enrichment analysis, and the exploration of protein functions and interaction networks within biological processes by employing specialized data processing tools such as Proteome Discoverer and MaxQuant, combined with continuously updated protein databases.
4. Experienced and Professional Technical Team
MtoZ Biolabs’ proteomics analysis service is supported by a team of experienced scientific experts with extensive project experience. This team is well-equipped to handle diverse and complex samples, ensuring scientifically sound experimental design and accurate data interpretation, ultimately driving research projects forward efficiently and effectively.
Applications
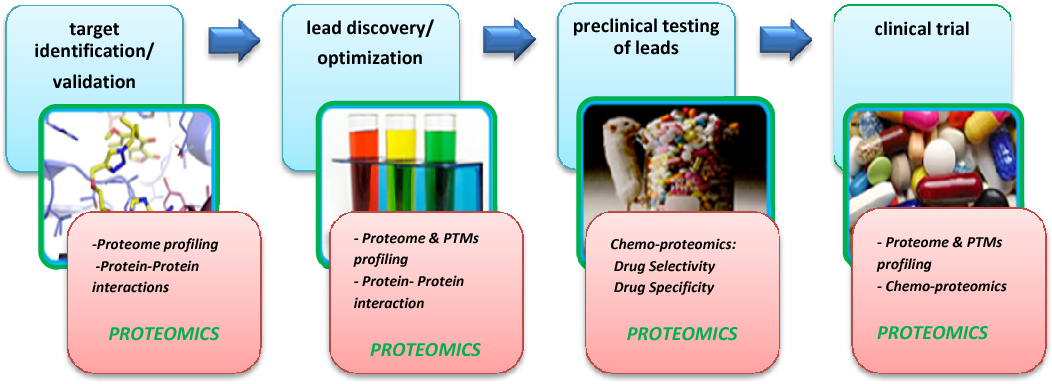
1. Target Identification and Validation: Proteomics analysis service enables protein expression profiling (Proteome Profiling) and protein-protein interaction studies (Protein-Protein Interactions) to identify and validate potential drug targets. These services provide insights into the biological functions of targets, laying a solid foundation for drug development.
2. Lead Compound Discovery and Optimization: At this stage, proteomics analysis service is utilized to combine post-translational modification (PTMs) analysis and protein interaction studies, offering a deeper understanding of the mechanisms of action between candidate compounds and their targets. This helps optimize the properties and efficacy of lead compounds, accelerating the drug development process.
3. Preclinical Testing of Leads: Proteomics analysis service applies chemo-proteomics approaches to evaluate the selectivity and specificity of drugs, providing comprehensive data support for the safety and efficacy of candidate compounds, and addressing critical challenges in drug development.
4. Clinical Trials: Combined with post-translational modification analysis and chemo-proteomicsproteomics analysis service is used to monitor patient drug responses, evaluate drug efficacy, and explore potential disease-related biomarkers, providing essential evidence for the implementation of precision medicine.
Nasrin, D. et al. Iran. J. Pharm. Sci. 2018.
Figure 10. Applications of Proteomics Analysis Service
Proteomics analysis service plays a pivotal role throughout the entire drug discovery and development pipeline, offering comprehensive and reliable solutions to advance new drug development and precision medicine.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Liquid Chromatography and Mass Spectrometry Parameters
4. The Detailed Information of Results
5. Mass Spectrometry Image
6. Raw Data
MtoZ Biolabs has much experience in proteomics analysis service with a comprehensive quality control workflow and experienced technical personnel capable of handling various types of samples. We can provide you with a one-stop proteomics analysis service and can tailor project plans based on actual research needs. You only need to tell us your experimental objectives and send us your samples, MtoZ Biolabs will take care of all subsequent matters. More analysis service is available for free consultation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







