Protein Deamidation Modification Analysis Service
Protein deamidation involves the conversion of amide groups (-CONH2) in amino acids like glutamic acid and aspartic acid into their corresponding acids, α-ketoglutarate and glutamic acid, by losing an ammonia molecule (NH3). This non-enzymatic post-translational modification is crucial for understanding proteins' charge distributions, functions, stability, and interactions with other molecules, as well as their roles in diseases. For example, β-amyloid proteins associated with Alzheimer's disease may become more prone to forming fibrils post-deamidation. Advances in molecular biology and mass spectrometry now allow for detailed analysis of such modifications. Using mass spectrometry, researchers can pinpoint exactly where deamidation occurs within protein sequences, which is essential for exploring protein functions, structures, and interactions.
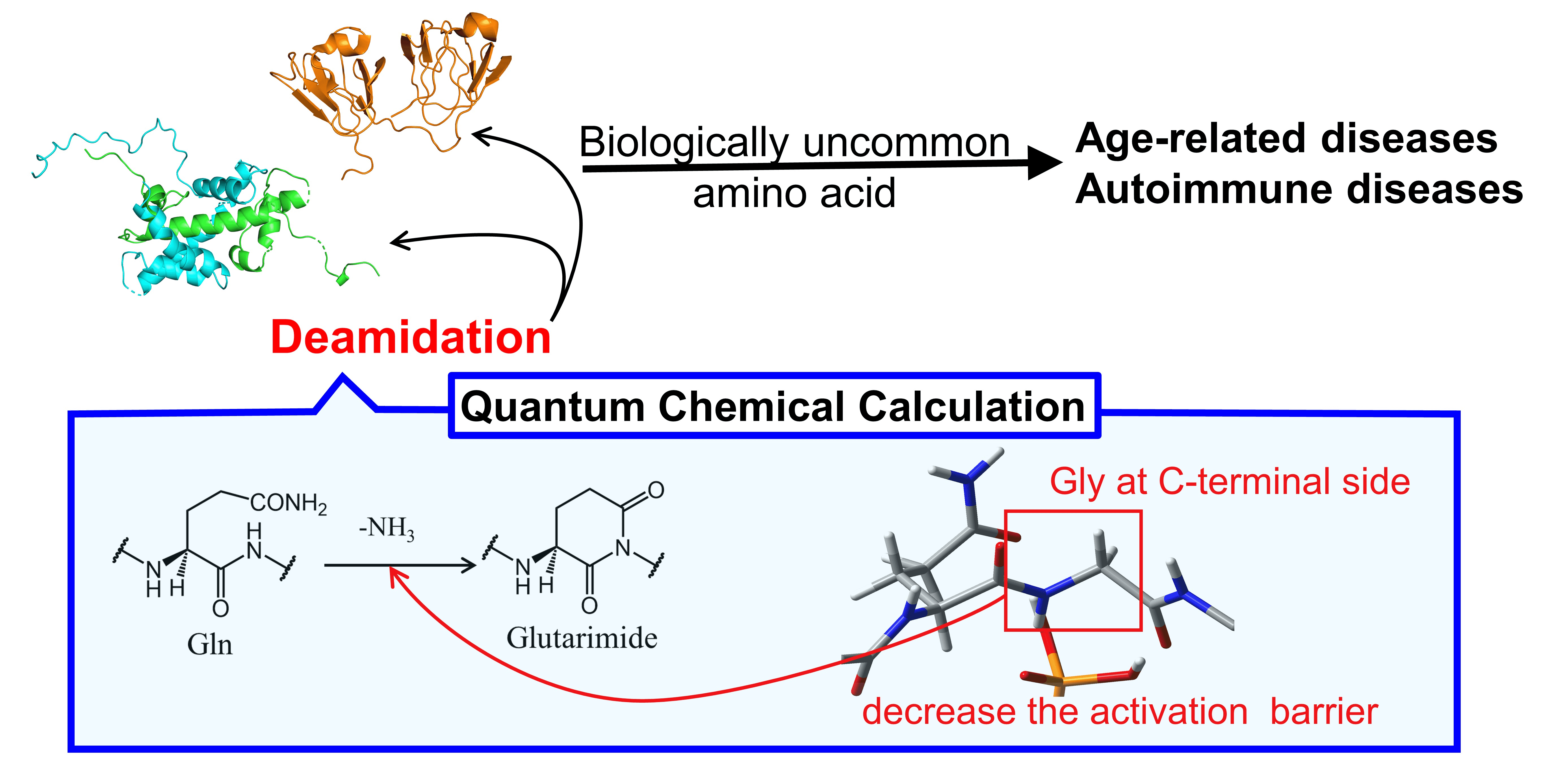
Asai, H. K. et al. AppliedChem. 2021.
Figure 1. Protein Deamidation Modification Process
Protein deamidation analysis via mass spectrometry is a sophisticated approach in protein research. This method involves inducing deamidation through specific enzymes or chemicals, converting aspartic and glutamic acids' carboxyl groups into amine groups. This change reduces the mass of these amino acids by about 1Da, creating distinct mass-to-charge ratios detectable in mass spectrometry. This differential allows for the precise identification and quantification of deamidation modifications in proteins. MtoZ Biolabs offers a high-resolution mass spectrometry-based platform for this analysis, ensuring high-quality, comprehensive services for studying cellular signaling, protein interactions, and disease mechanisms.
Deliverables
In the technical report, MtoZ Biolabs will provide you with detailed technical information, including:
1. Experimental Procedures
2. Relevant Experimental Parameters
3. Detailed Information on Protein Deamidation Modifications
4. Mass Spectrometry Images
5. Raw Data
How to order?







