Antibody Sequence Analysis Service
Antibody Sequence Analysis Service generally refers to determining the amino acid sequence of antibody molecules using high-resolution mass spectrometry and other advanced tools. The sequence of antibody molecules, particularly the amino acid sequence of the variable region, is central to the antibody's biological function. Accurate analysis of antibody sequences is essential for understanding the structural and functional characteristics of antibodies. Antibody sequence analysis can comprehensively analyze the sequences of antibody heavy chain (HC) and light chain (LC), and identify post-translational modifications (PTMs), such as glycosylation, oxidation, and deamination. Using high-resolution mass spectrometers, MtoZ Biolabs offers precise antibody sequence analysis service for different antibody subtypes (e.g., IgG and IgM) and various antibody types (e.g., fluorescent-labeled antibodies, immobilized antibodies, and antibodies from different species). Our service can analyze both the full-length sequence and the variable region sequence of antibodies, providing complete sequence information for LC and HC.
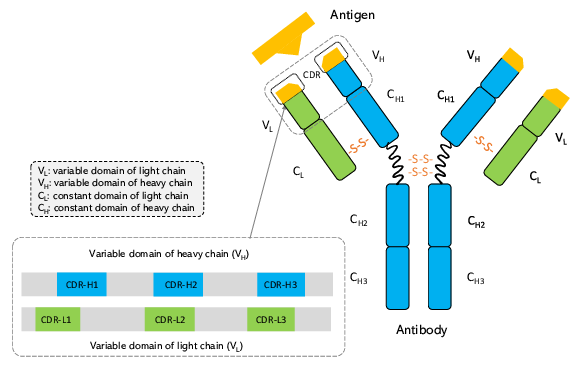
Gao K, et al. Proceedings of the 29th KDD. 2023.
Analysis Workflow
The primary workflow of antibody sequence analysis includes:
1. Antibody Sample Digestion
The antibody sample is digested using specific proteases to produce detectable peptide fragments.
2. High-Resolution Mass Spectrometry (MS) Detection
Peptide fragments are analyzed using a high-resolution mass spectrometer to obtain precise molecular weight and sequence fragment information.
3. Database Matching and Assembly
The acquired peptide fragment data is matched against an antibody sequence database to assemble the complete antibody sequence.
4. Result Validation
Bioinformatics algorithms and secondary validation techniques are applied to ensure the accuracy and completeness of the sequence.
Applications
Antibody sequence analysis has many applications in biopharmaceuticals, immunology, and fundamental scientific research, including:
Antibody Drug Development
Use antibody sequence analysis to ensure antibody consistency and reproducibility during monoclonal antibody (mAb) drug development.
Basic Immunology Research
Use antibody sequence analysis to identify specific autoantibodies in autoimmune disease studies, offering new targets for disease diagnosis and treatment.
Antibody Engineering
During the process of antibody humanization, key antibody regions are identified through antibody sequence analysis and comparison, and targeted modifications are performed to reduce the immune response.
Biomarker Discovery
By analyzing the antibody sequences in patient serum or tissues, antibody molecules associated with specific diseases can be identified.
FAQ
Q: During the antibody sequence analysis, how to ensure that the sequences of the HC and LC of the antibody are fully covered? Especially for the variable region, how to avoid the loss of sequence information at key sites?
Ensuring sequence integrity and coverage is one of the most fundamental and critical challenges in antibody sequence analysis. Sequence completeness directly affects the accuracy of the final results. In the analysis of the variable region, even a single missing key amino acid could lead to misinterpretation of the antibody's binding properties.
1. Multi-Enzyme Digestion Strategy
To ensure sequence coverage, a multi-enzyme digestion strategy can be employed. Different proteases target specific amino acid residues, producing peptide fragments of varying lengths and positions, thereby minimizing blind spots in the sequence and improving overall sequence coverage.
2. High-Resolution Mass Spectrometry Detection
Using a high-resolution mass spectrometer (such as Orbitrap Fusion Lumos or Q Exactive HF) can significantly improve the sensitivity and resolution in peptide detection, reduce the risk of missing low-abundance peptides. Additionally, combining fragmentation techniques like CID (Collision-Induced Dissociation), HCD (Higher-energy Collisional Dissociation), and ETD (Electron Transfer Dissociation) can ensure efficient fragmentation and detection of diverse peptide types.
3. Database Matching and Assembly Optimization
Use high-quality antibody sequence databases such as IMGT or NCBI for multiple rounds of database matching to ensure sequence integrity. At the same time, use bioinformatics assembly algorithms, such as Byonic and PEAKS, to accurately splice and align sequences.
4. Sequence Blind Spot Validation
For regions difficult to detect via mass spectrometry, nucleic acid sequencing (such as cDNA sequencing) can be used for supplementary verification to make up for the limitations of mass spectrometry detection. In addition, immunological verification methods such as Western Blot can be combined to ensure the accuracy of key sites.
5. Internal Standards and Quality Control Samples
Standard antibodies are introduced as internal controls during the analysis process to monitor the coverage and integrity of the experiment in real time to ensure the controllability and repeatability of the experimental process.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
Researchers performed antibody sequence analysis on polyclonal IgG derived from human plasma, and successfully identified and generated 12 recombinant antibodies. Some of which exhibited binding affinities comparable to or even higher than those of the natural polyclonal antibodies.
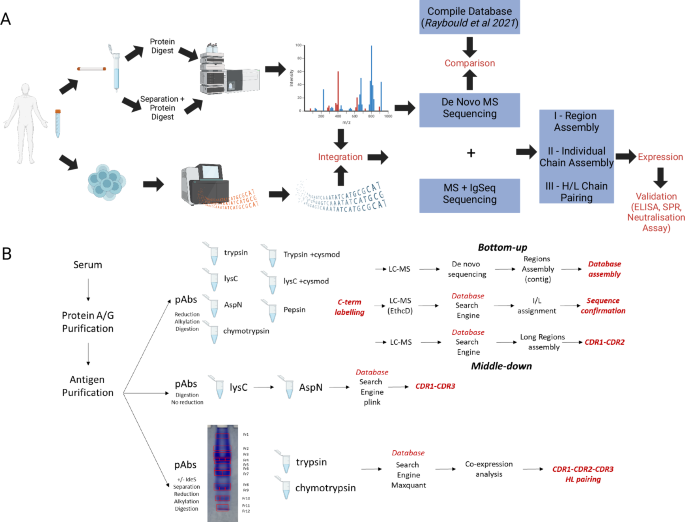
Bihan T L. et al. Nature Communications. 2024
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs provides mass spectrometry-based antibody sequence analysis service and PCR-based monoclonal antibody sequencing service. Feel free to contact us for more information.
How to order?







