Blood Proteomics Service
Blood proteomics is the science of analyzing the protein composition of blood to study their functions within the organism and their interactions with other biomolecules, encompassing both plasma proteomics and serum proteomics. Proteins are the most critical macromolecules in blood samples, acting as the executors of biological activities and functions. However, the complexity of blood samples presents significant challenges for proteomics research.
Analyzing blood samples in proteomics is particularly challenging because high-abundance proteins, such as albumin and IgG, constitute over 90% of the total protein content. Mass spectrometry (MS) often repeatedly scans these abundant proteins, making it difficult to detect meaningful low-abundance proteins. Consequently, conventional strategies often require the removal of high-abundance proteins and pre-fractionation, which can result in the loss of important protein information in the blood.
MtoZ Biolabs offers a comprehensive solution to these challenges in blood proteomics through its multifunctional and powerful data-independent acquisition (DIA) platform. This platform combines the latest advancements in MS with MtoZ Biolabs' expertise in proteomics software and services. It is user-friendly and scalable, making it an ideal choice for researchers of all experience levels.
Technical Principles
At the heart of blood proteomics technology lies MS. This technique transforms protein molecules into charged ions using an ionization source, then separates and detects these ions with the aid of electromagnetic fields. By analyzing the mass-to-charge ratio (m/z) and abundance of the ions, MS enables the identification and quantification of proteins.
Analysis Workflow
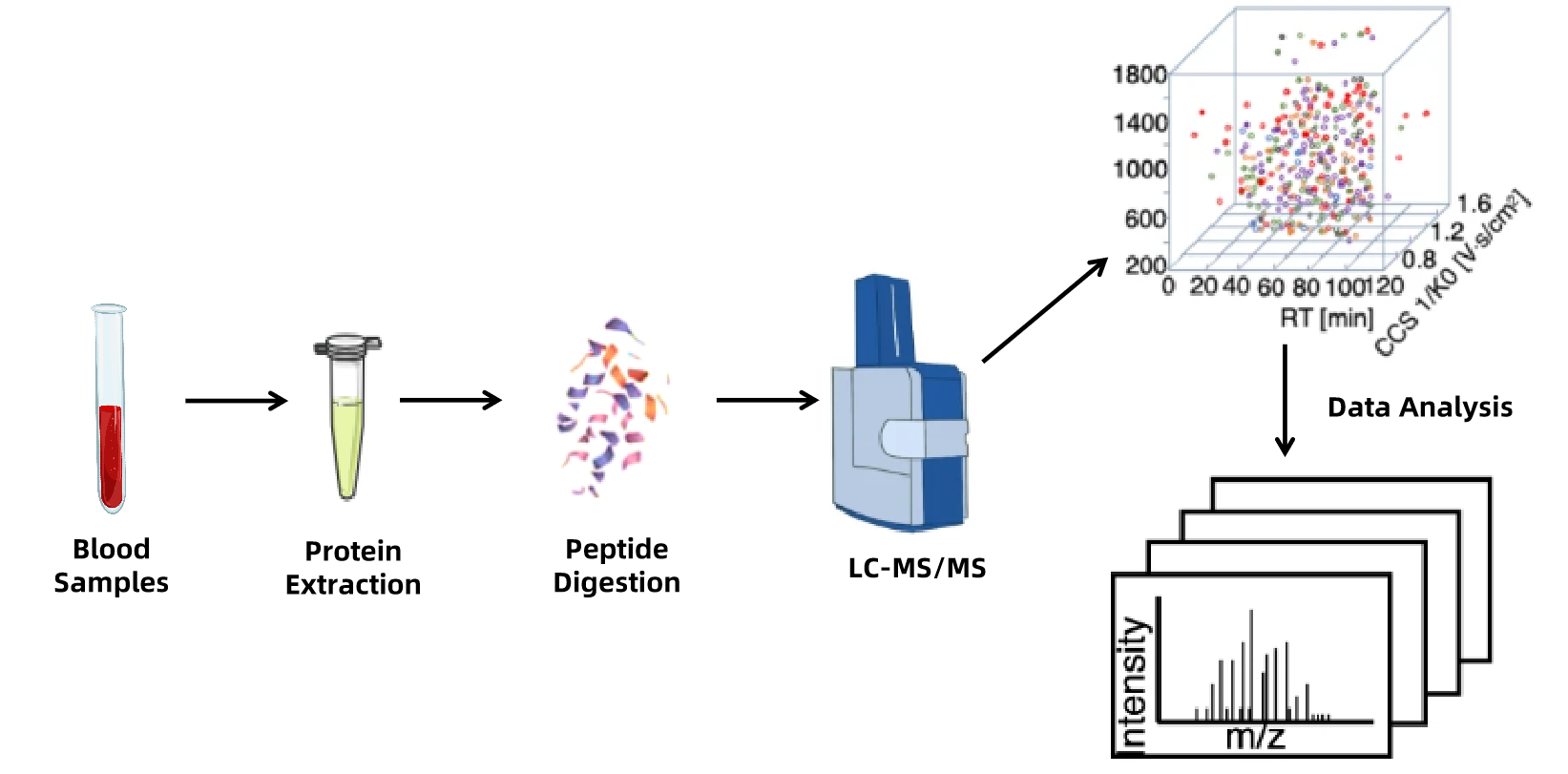
Figure 1. Blood Proteomics Workflow
Service Advantages
1. High Reliability
Utilizing the DIA full-scan data acquisition mode reduces interference from high-abundance signals, enabling accurate identification of low-abundance proteins.
2. High Throughput
Capable of quantifying thousands of proteins within a sample simultaneously. Under identical analysis conditions, its analytical capacity is more than double that of traditional proteomics techniques (such as iTRAQ, TMT, or label-free methods), making it more suitable for large-scale sample analyses.
3. Non-invasive and Reproducible
Blood sample collection is convenient and repeatable, making blood proteomics an ideal tool for disease screening, diagnosis, and monitoring.
4. Wide Applicability
This technology is suitable for various biological samples and experimental conditions, offering broad applicability and can be integrated with other techniques to form a multi-omics analysis platform.
Sample Submission Requirements
Sample: Not less than 1 mL of blood
Applications

Figure 2. Applications of MS-based Proteomics in Hematology and Blood Diseases
Sample Results
Proteomics of Prostate Cancer in Peripheral Blood Identified Using DIA-MS Combined with Machine Learning
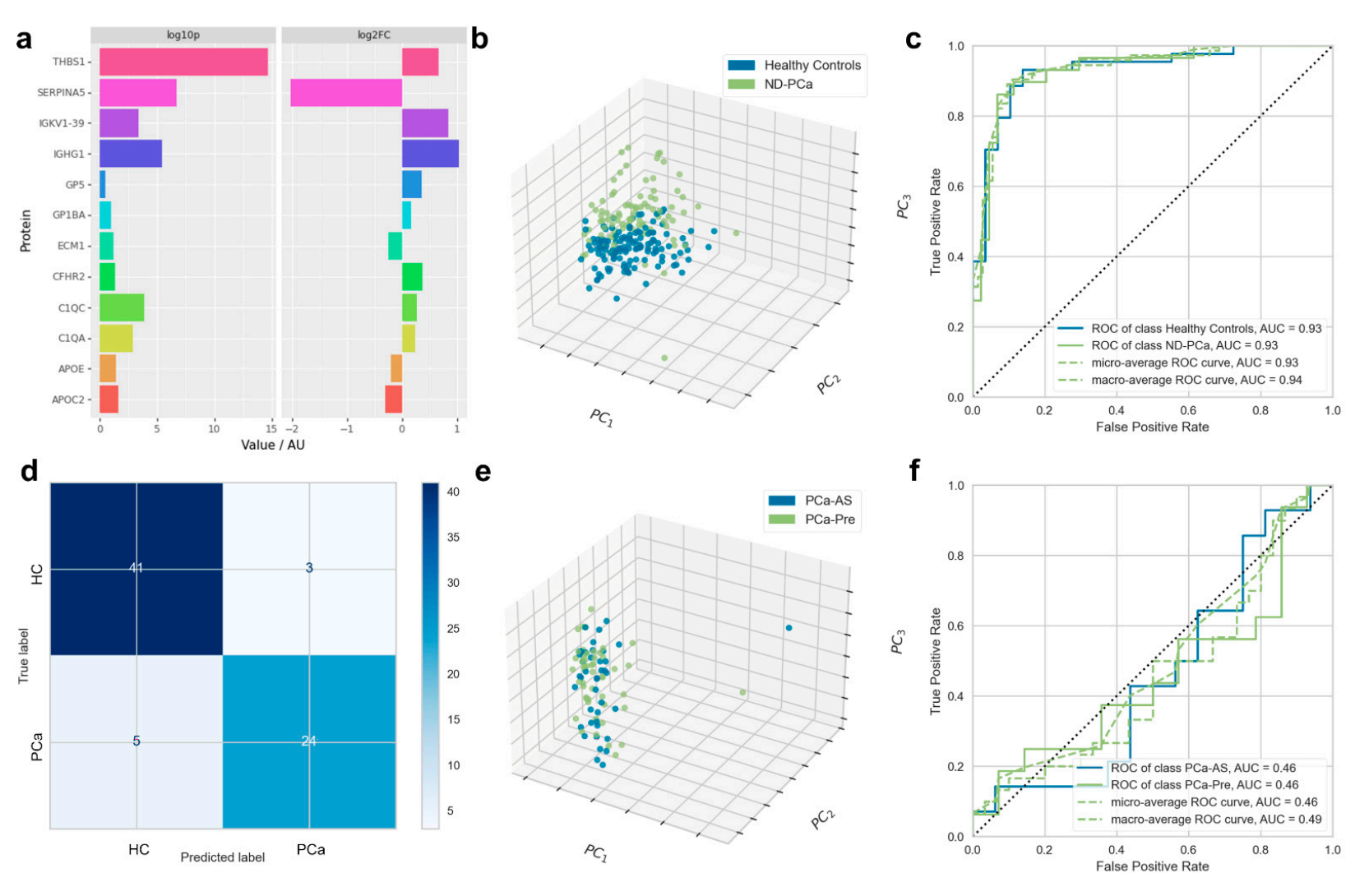
Muazzam, A. et al. Cancers. 2023.
Deliverables
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Detailed Information on Blood Proteomics
4. Mass Spectrometry Images
5. Raw Data
FAQ
Q1: Whether serum or plasma is better for proteomics?
In proteomics research, the choice between using serum or plasma depends on the research objectives and experimental design. Serum, with its high stability and straightforward processing, is suitable for studies requiring high stability, such as long-term health monitoring and chronic disease research. Plasma, on the other hand, offers a more comprehensive protein profile, making it ideal for studies that need to reflect dynamic changes within the body, such as acute disease and inflammatory response research. However, the anticoagulants in plasma may interfere with analysis, and its processing steps are more complex.
Q2: What are the requirements for proteomic blood samples?
Both plasma proteomics and serum proteomics require starting with blood to obtain plasma or serum. Plasma is obtained by adding an anticoagulant to the blood and then centrifuging it to collect the supernatant. Serum is obtained by allowing the blood to clot and then centrifuging it to collect the supernatant. Researchers should process the blood based on their specific research objectives. If mass spectrometry analysis is to be used subsequently, EDTA is not recommended as an anticoagulant. The blood samples should be stored at -80°C and should not undergo repeated freeze-thaw cycles.
How to order?







