Chemical Proteomics: Precision Dissection of Protein Mechanisms, Opening New Frontiers in Scientific Research
Chemical proteomics is an emerging interdisciplinary field that integrates chemical techniques with proteomic analysis, aiming to selectively label, separate, and identify specific proteins in complex biological systems using chemical methods. This technology can reveal the structure and function of proteins within living organisms and has significant potential in drug discovery, disease diagnosis and treatment, as well as basic biological research. Through chemical proteomics, scientists can more efficiently identify and analyze disease-related protein targets, providing more reliable foundations for the precise design of drugs and advancing our understanding of complex molecular mechanisms in living organisms. This integrative approach is becoming an essential tool in modern life sciences research.
As a key player in this field, MtoZ Biolabs is dedicated to providing efficient and accurate chemical proteomics services to researchers. With advanced mass spectrometry platforms, mature chemical labeling technologies, and a professional research team, MtoZ Biolabs excels in selectively labeling, separating, and accurately identifying specific proteins within complex biological systems. This helps reveal key protein molecular mechanisms and their roles in biological processes. Below are some specific applications of chemical proteomics.
ABPP (Activity-based Protein Profiling) / CCCP (Compound-Centric Chemical Proteomics)
In chemical proteomics, Activity-based Protein Profiling (ABPP) and Compound-Centric Chemical Proteomics (CCCP) are core techniques that leverage the specific affinity between small molecules and proteins to isolate and identify target proteins. These methods involve designing small molecule probes with specific chemical structures that maintain biological activity, while incorporating reactive groups, affinity tags, and reporter groups to efficiently and precisely label, separate, and detect target proteins. Subsequent proteomic analyses of these labeled proteins provide insights into their structure, function, and interaction with small molecules within biological systems. These approaches offer powerful tools for drug target discovery and validation, disease-related protein identification, and the exploration of biological mechanisms, thereby accelerating the application of chemical proteomics in life sciences.
1. Advantages
The ability to specifically label active proteins in situ within complex biological systems.
2. Limitations
Probes must be synthesized. Most initial operations, such as probe design, validation, and preliminary protein labeling and analysis, are performed in vitro. The in vitro conditions may differ from the true physiological environment in vivo (e.g., pH, ion concentration, and regulatory factors).
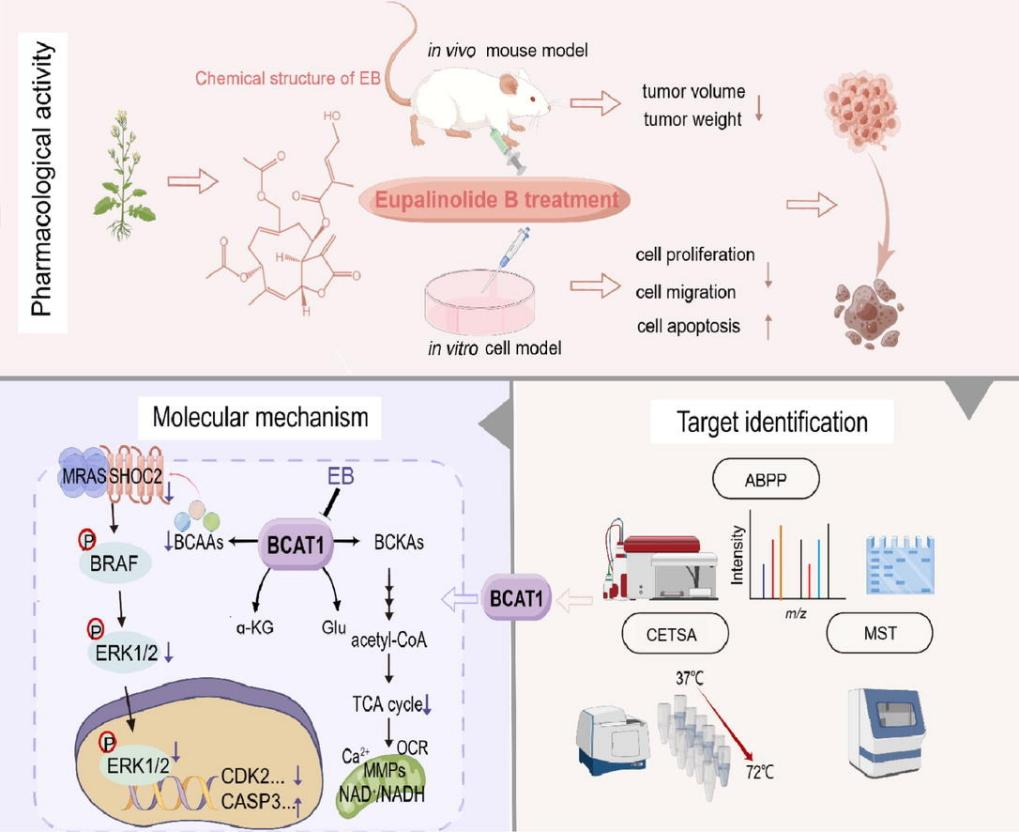
Figure 1.
A recent study (2024) titled "Small-molecule targeting BCAT1-mediated BCAA metabolism inhibits the activation of SHOC2-RAS-ERK to induce apoptosis of triple-negative breast cancer cells" reported that EB-P (wild horse chase lactone B), a naturally derived small-molecule probe, directly targets the Cys335 residue of BCAT1. This interaction significantly inhibits BCAT1-mediated BCAA metabolism and suppresses the activation of the SHOC2-RAS-ERK signaling pathway, ultimately leading to apoptosis in triple-negative breast cancer (TNBC) cells.
MS-based Thermal Stability Profiling / In-cell Thermal Shift Analysis
In the field of chemical proteomics, MS-based thermal stability profiling and in-cell thermal shift analysis are widely adopted to study changes in protein thermal stability before and after their interaction with small molecules. These techniques exploit the principle that ligand binding often alters the denaturation temperature of a protein. By gradually increasing the temperature and combining it with quantitative mass spectrometry, researchers can monitor protein solubility changes in real time, allowing for the identification of potential target engagement events in living cells or lysates. This approach enables the unbiased identification of drug–protein interactions across the proteome under near-native conditions, providing mechanistic insights into drug action, as well as supporting target deconvolution and drug screening efforts.
1. Advantages
This method allows for unbiased identification of drug protein targets in their native environment and across the entire proteome.
2. Limitations
Proteins with weak binding affinities may show minimal changes in thermal stability, making them difficult to detect. Furthermore, it cannot distinguish between direct interactions between drugs and proteins and indirect interactions mediated by other molecules.
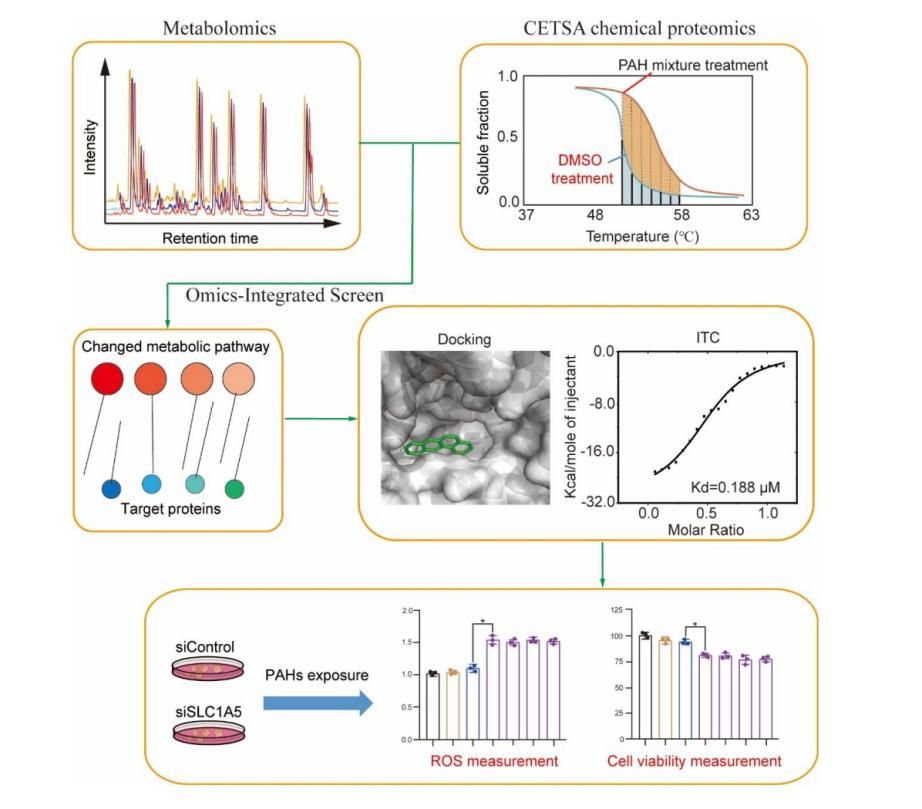
Figure 2.
A recent study (2024), titled "Benzo(a)anthracene Targeting SLC1A5 to Synergistically Enhance PAH Mixture Toxicity", identified that BaA (benzo[a]anthracene) targets the SLC1A5 protein to enhance oxidative damage caused by PAH mixtures, thereby amplifying their synergistic toxic effects. The study employed metabolomics, MS-based thermal stability profiling, molecular docking, and isothermal titration calorimetry for this discovery.
LIP-MS (Limited Proteolysis-Mass Spectrometry) / DARTS (Drug Affinity Responsive Target Stability)
In the field of chemical proteomics, Limited Proteolysis-Mass Spectrometry (LIP-MS) and Drug Affinity Responsive Target Stability (DARTS) are two techniques based on differences in the resistance of proteins to proteases. The core principle of these techniques is that when a protein's conformation changes (e.g., upon binding to a small molecule), its protease cleavage sites may become shielded or exposed due to changes in steric hindrance, leading to significant alterations in its resistance to proteases. In practice, these techniques achieve limited proteolysis by preferentially cleaving peptide regions that are surface-exposed, flexible, or interacting with other molecules. These peptides are then analyzed using high-resolution mass spectrometry, allowing for the inference of structural changes, interaction regions, and functional alterations of the protein upon binding with small molecules. This strategy not only aids in elucidating drug binding sites on target proteins, but also uncovers potential drug mechanisms of action and protein dynamic conformational changes, providing a powerful tool for chemical proteomics in drug target identification and functional protein research.
1. Advantages
Provides structural and functional information about proteins, capturing their dynamic changes. It allows for the detection of protein alterations induced by various factors in their native state without requiring extensive purification or special treatments.
2. Limitations
This method cannot determine the specific binding sites of drugs on proteins, the affinity between the drug and the protein, or the precise effects of their interaction on protein function.
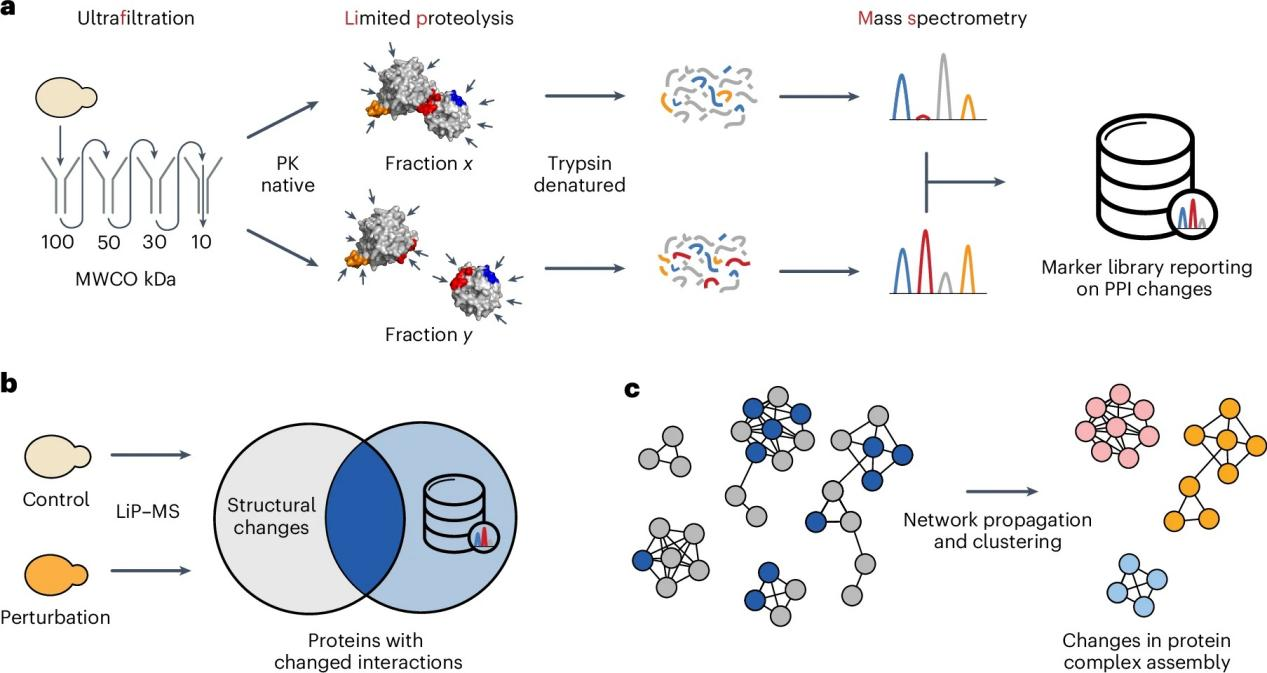
Figure 3.
On October 16, 2024, the research team led by Professor Paola Picotti at the Swiss Federal Institute of Technology in Zurich (ETH Zurich) combined ultrafiltration with FLiP-MS to analyze the proteome of Saccharomyces cerevisiae. They identified protein-protein interaction (PPI) markers for 1,086 proteins and assessed these candidates through experimental data, AlphaFold-predicted structures, functional enrichment analysis, and known mutations affecting PPIs. The identified PPI markers were subsequently employed to track global protein dynamics in yeast cells under hydroxyurea treatment.
PROTAC (Proteolysis-Targeting Chimeras)
In the field of chemical proteomics, PROTAC (Proteolysis-Targeting Chimeras) is an innovative small-molecule technology designed to achieve efficient degradation of target proteins through specifically engineered small-molecule chimeras. These chimeras consist of two functional parts: one end specifically binds to the target protein, while the other binds to the E3 ubiquitin ligase. This bifunctional molecule bridges the target protein and the E3 ligase, promoting ubiquitination of the target protein, which is subsequently recognized and degraded by the proteasome.
Unlike traditional small-molecule inhibitors, PROTAC not only inhibits protein activity but also completely degrades specific target proteins, helping to avoid off-target effects that inhibitors may cause. This technology offers a novel approach to druggable protein targets that were previously considered undruggable, showing significant potential in the treatment of cancer, neurodegenerative diseases, and other conditions related to protein dysfunction. As a tool in chemical proteomics, PROTAC technology opens new research directions for drug discovery and targeted therapy.
1. Advantages
Rather than relying on inhibiting or activating the active site of a protein, PROTAC induces the binding of the target protein to the ubiquitin ligase, leading to its ubiquitination and subsequent degradation by the proteasome. This enables intervention in traditionally “undruggable” targets, greatly expanding the range of druggable targets in drug development.
2. Limitations
PROTAC molecules are relatively large and structurally complex, making them difficult to enter cells or be metabolized. Additionally, they can cause off-target effects, and their clinical application still requires further refinement.
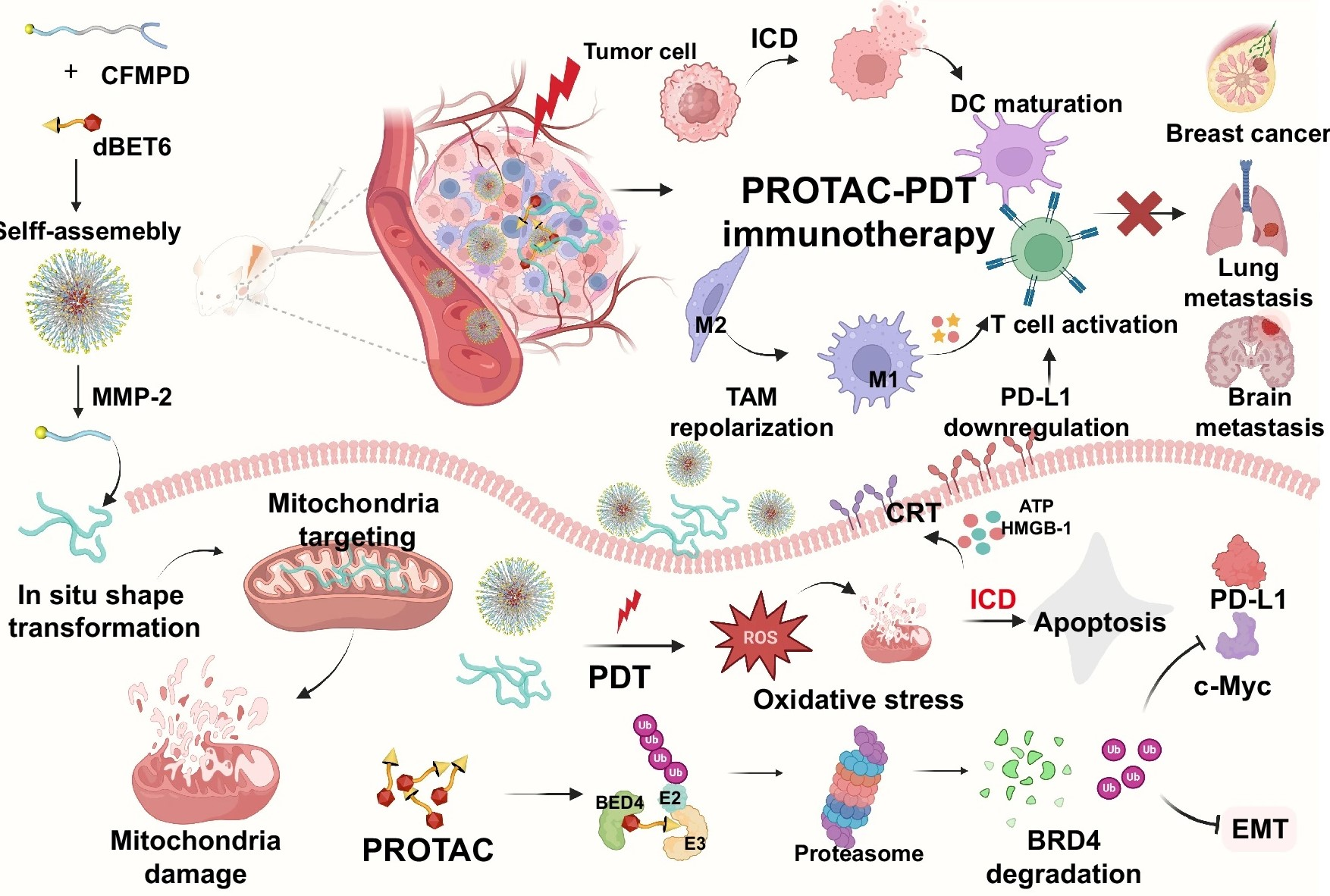
Figure 4.
A study published in November 2024, titled "MMP-2-triggered, mitochondria-targeted PROTAC-PDT therapy of breast cancer and brain metastases inhibition", investigated the integration of PROTAC technology with photodynamic therapy (PDT) and mitochondrial targeting. The findings revealed that PROTAC-PDT molecules were effectively absorbed by both breast cancer cells and brain metastasis cells. Triggered by MMP-2, these molecules selectively released active PROTAC and photosensitizers, thereby enabling targeted and precise tumor cell therapy.
Chemical proteomics, as an advanced discipline integrating chemical tools with proteomics technologies, has gradually become a powerful means of revealing protein functions, analyzing drug targets, and exploring biological mechanisms. From ABPP/CCCP affinity screening, MS-based thermal stability profiling/in-cell thermal shift analysis, to LIP-MS/DARTS proteolytic resistance differences analysis, and PROTAC targeted degradation strategies, these techniques together form a comprehensive and efficient protein research toolkit, providing robust technical support for drug development, disease diagnosis, and fundamental biological research. As a pioneer in the field of biological mass spectrometry, MtoZ Biolabs has a professional technical team. We offer comprehensive and efficient chemical proteomics services. By choosing MtoZ Biolabs, you will benefit from expert technical support, precise data analysis, and customized research solutions.
References
[1] DOl: 10.1016/j.jare.2024.10.021
[2] DOl: 10.1021/acs.est.4c07053
[3] DOl: 10.1038/s41587-024-02432-8
[4] DOI: 10.1038/s41467-024-54854-2
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







