CUT&Tag Analysis Service
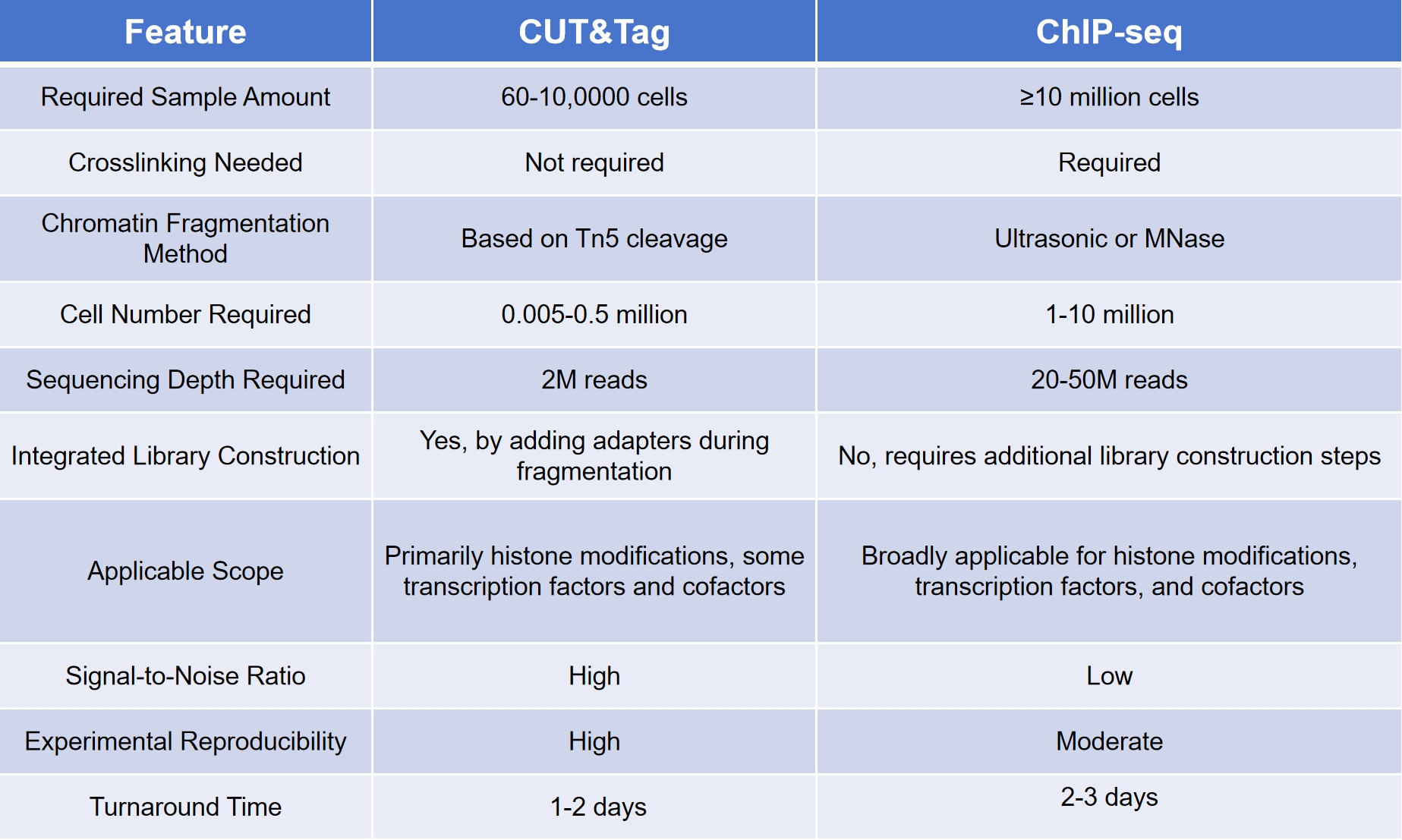
- Lower background noise: Eliminates the need for chemical crosslinking, reducing nonspecific binding.
- Higher resolution: Enables precise localization of transcription factor and histone modification binding regions.
- Optimized timeline: Integrates fragmentation and library construction processes, significantly reducing the experimental timeline.
- Use cryopreservation medium consisting of 10% DMSO + 90% fetal bovine serum and maintain transport at 4°C.
- Alternatively, flash-freeze the samples in liquid nitrogen and store at temperatures below -80°C, transporting with dry ice.
CUT&Tag (Cleavage Under Targets and Tagmentation) is an innovative tool dedicated to DNA-protein interaction research, primarily used to identify transcription factor or histone modification binding sites on a genome-wide scale. The CUT&Tag analysis service utilizes cleavage and transposase tagging technology to precisely map the binding sites of transcription factors or histone modifications within the genome, facilitating research into DNA-protein interaction mechanisms. It provides an accurate solution for genomic and epigenetic studies. Its high sensitivity, low background noise, and exceptional adaptability to low-input samples make it stand out in the field of epigenetics research.
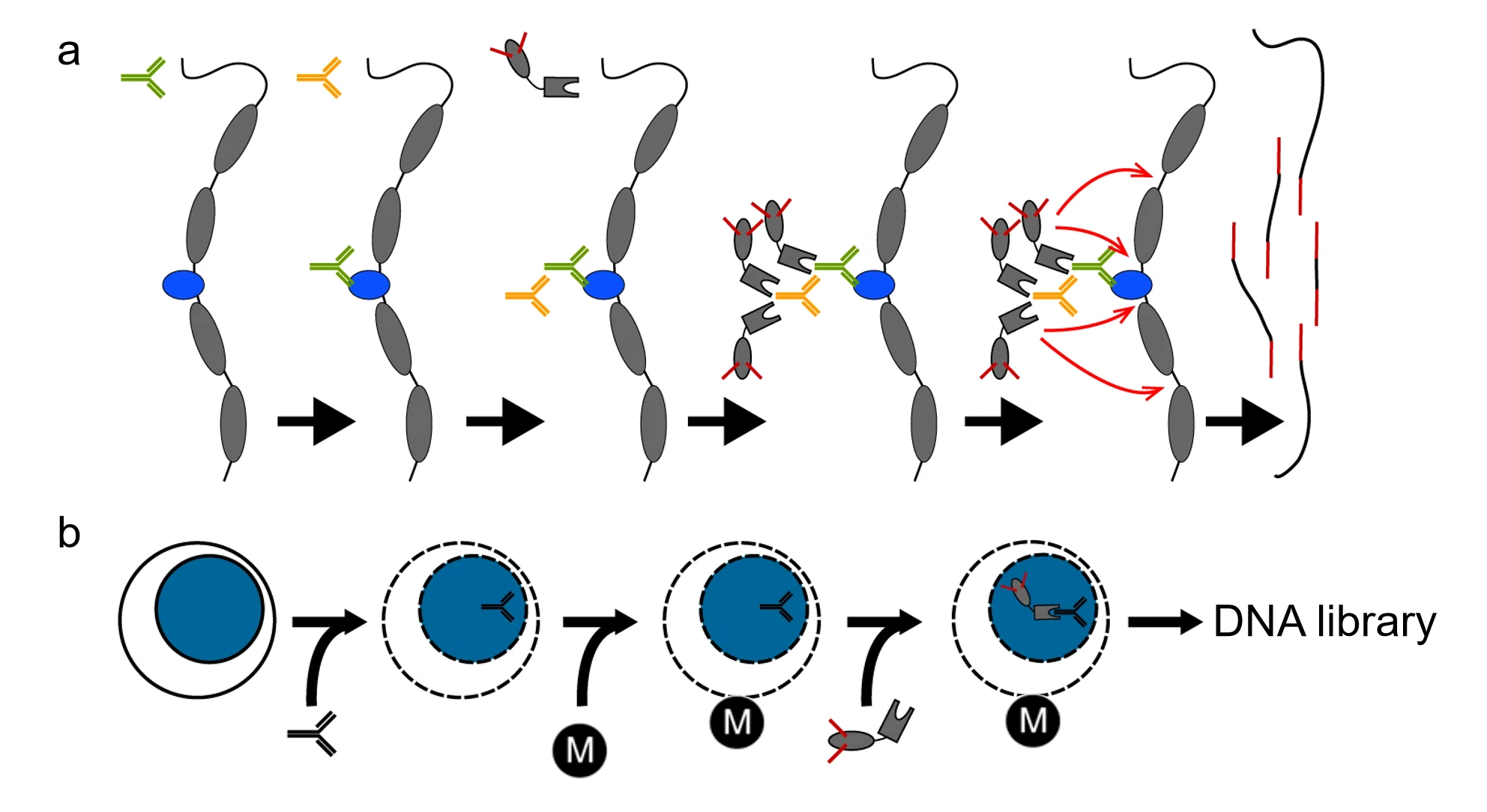
Kaya-Okur, H.S. et al. Nat Commun. 2019.
Principles of CUT&Tag Analysis
In genomic and epigenetic studies, exploring the mechanisms of DNA-protein interactions is key to understanding gene regulation. Transcription factors regulate the recruitment of RNA polymerase and the initiation of gene transcription by binding to specific DNA sequences in promoter or enhancer regions. The efficiency of their binding determines the spatiotemporal pattern of gene expression. Histones, as the core components of chromatin, regulate chromatin accessibility and DNA availability through post-translational modifications (e.g., acetylation and methylation), thereby influencing the binding of transcription factors to DNA. Such interactions can either activate gene expression or induce silencing. Protein-DNA interaction analysis elucidates the synergy between transcription factors and histone modifications in gene regulation, providing crucial insights into epigenetic mechanisms and disease pathogenesis.
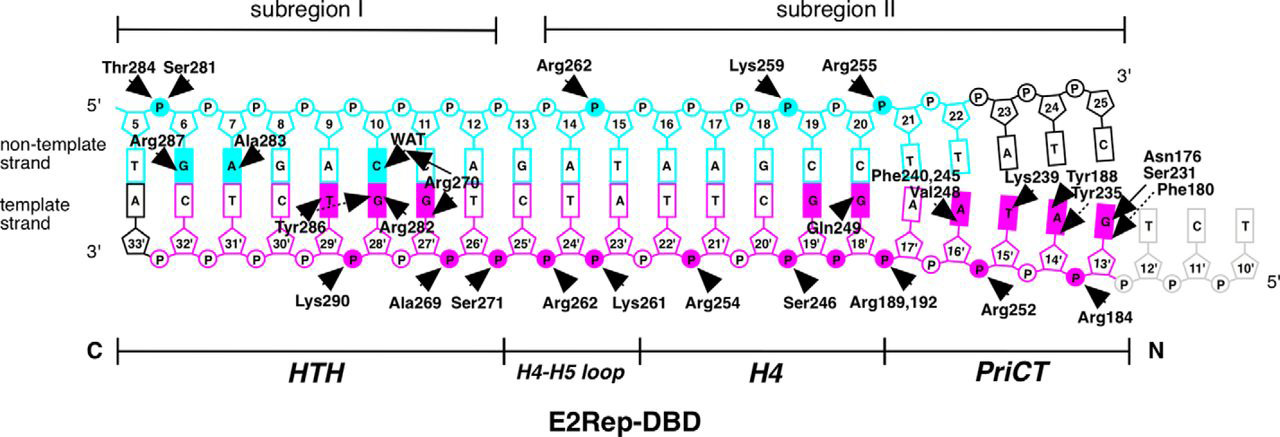
Itou H,et al.J Biol Chem.2015.
Schematic Diagram of the Protein-DNA Interactions Observed in the Crystal Structure
CUT&Tag employs Protein A/G-Tn5 fusion proteins to directly bind target proteins, eliminating the need for traditional crosslinking steps, thereby significantly reducing background noise and achieving high-resolution detection of binding sites. Compared to traditional ChIP-seq, CUT&Tag is more suitable for low-input samples, particularly excelling in studies of histone modifications and transcription factor binding sites. It serves as a powerful tool for uncovering gene regulatory mechanisms, offering efficient solutions for disease research and drug development.
Service at MtoZ Biolabs
MtoZ Biolabs provides the CUT&Tag analysis service leveraging an advanced Protein A/G-Tn5 fusion protein system and high-throughput sequencing platforms. We specialize in analyzing protein-DNA interactions, with a focus on transcription factor binding sites and the genome-wide distribution of histone modifications. Our CUT&Tag analysis service encompasses the entire workflow including sample preparation, sequencing, and data analysis while offering comprehensive genome annotation, functional analysis, and pathway mapping. We empower clients to unravel gene regulatory networks with precision, advancing epigenetics, disease research, and drug discovery.
1. Sample Preparation and Bead Binding
ConA beads are used to bind glycoproteins on the cell membrane, allowing for the extraction of nuclei or the direct use of nuclear samples. Simultaneously, digitonin is applied to permeabilize the cell membrane, enhancing nuclear membrane permeability and facilitating subsequent antibody binding.
2. Primary and Secondary Antibody Binding
Specific antibodies (primary antibodies) targeting the protein of interest are added and incubated (e.g., α-H3K27me3 as a positive control and isotype control IgG as a negative control), followed by washing to remove nonspecific bindings. Secondary antibodies are then added to ensure successful binding of the antibody complex to the target region.
3. Protein A/G-Tn5 Transposome Binding
Protein A/G-Tn5 fusion protein is used to bind the antibody complex, precisely directing the Tn5 transposase to the DNA regions associated with the target protein. This creates a highly efficient transposome complex, laying the foundation for subsequent fragmentation and adapter integration.
4. Tn5 Transposase Activation and DNA Fragmentation
A reaction buffer containing Mg²⁺ is added to activate the Tn5 transposase, enabling precise fragmentation of DNA at the target binding sites while simultaneously inserting sequencing adapters into the DNA fragments, completing the initial library construction.
5. Sequencing and Bioinformatics Analysis
High-throughput sequencing of the library is performed using Illumina platforms to obtain high-resolution data. Professional data analysis platforms are employed for quality control, genome alignment, peak annotation, motif identification, GO functional analysis, and KEGG pathway analysis, providing comprehensive insights into the functional roles of transcription factors and histone modifications.
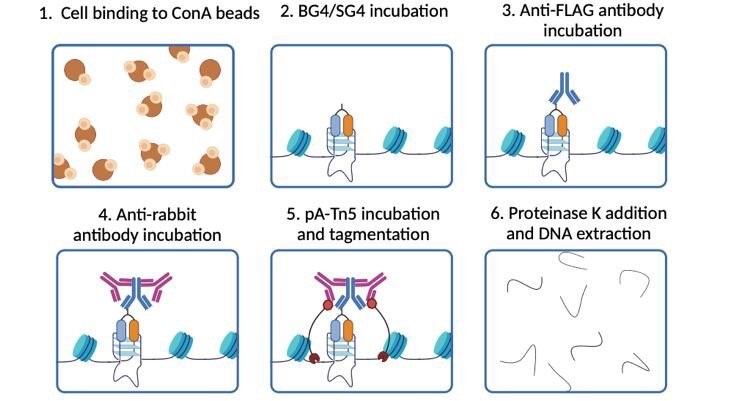
Galli, S. et al. RSC Chem Biol. 2024
CUT&Tag Analysis Workflow
Technical Advantages
Compared to traditional ChIP-seq, CUT&Tag excels across multiple dimensions:
Sample Submission Suggestions
MtoZ Biolabs’ CUT&Tag analysis service employs efficient transposase tagging and cleavage technologies to provide precise protein-DNA interaction analysis for genomic and epigenetic research. Proper sample preparation and storage are key to achieving high-quality analysis. We recommend handling your samples as follows:
Cell Samples
1. Cleaning and Digestion: Remove the old culture medium, wash the cells once with PBS, and add an appropriate amount of trypsin to gently digest the cell layer.
2. Centrifugation and Washing: Centrifuge at 1000 rpm for 5 minutes, discard the supernatant, and wash the cells once more with PBS.
3. Storage Preparation: Resuspend the cells in an appropriate volume of pre-prepared cryopreservation medium, gently pipette to mix, and count the cells. Each tube should contain at least 500,000 cells.
4. Aliquot and Freeze: Aliquot the cells into cryotubes, adding 150 μl of cryopreservation medium per tube. Ensure each sample has 2-3 parallel tubes. Store the samples in a pre-cooled -80°C freezer and ship using dry ice.
Tissue Samples
1. Sampling and Cleaning: Fresh tissues obtained from live specimens should be washed with PBS to remove surface impurities. Temporarily store the samples in 1640 or DMEM medium at 4°C, but process them within 3 hours.
2. Cutting and Processing: Cut the tissues into small pieces or mince them into a paste-like form, and aliquot into multiple cryotubes, ensuring 2-3 tubes per sample.
3. Cryopreservation Options:
For other special sample types, please feel free to contact us.
Sample Results
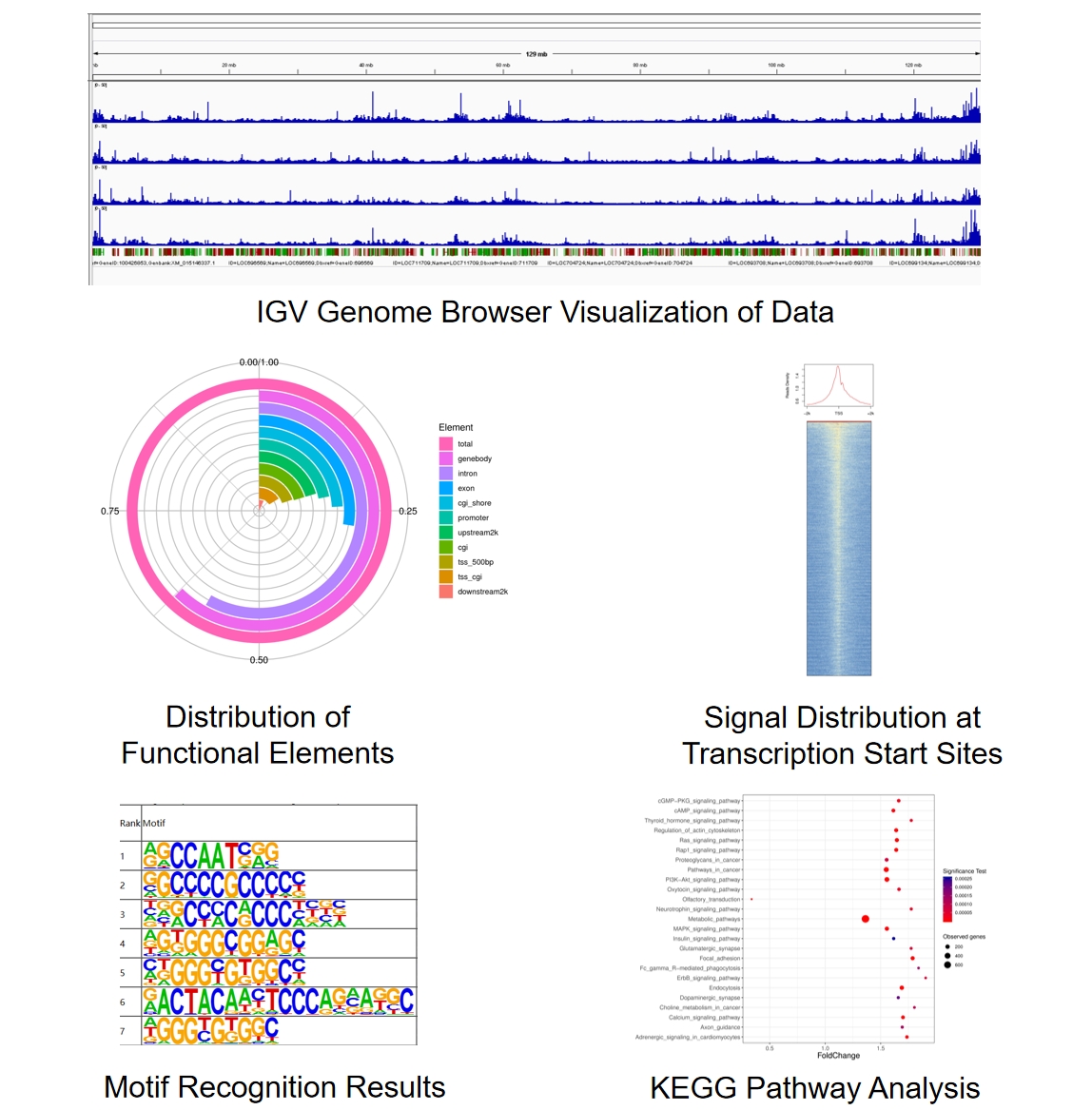
Case Study
CUT&Tag Reveals the Genomic Distribution and Gene Expression Impact of Histone Lactylation in Pancreatic Ductal Adenocarcinoma (PDAC)
CUT&Tag technology was used to investigate the genomic distribution of lactylation modifications in PDAC and their impact on gene expression. CUT&Tag was employed to map histone lactylation (H3K18la) binding patterns in the genome of pancreatic cancer cells. Results showed that H3K18la was primarily enriched at promoters and enhancers of tumor-related genes. RNA-seq data further revealed that lactylation levels in these regions were closely associated with the high expression of glycolytic and tumor-related genes.
The study demonstrated that lactate produced from glycolytic metabolism drives histone lactylation, which in turn promotes the expression of key glycolytic genes, forming a positive feedback loop that accelerates tumor progression. This mechanism underscores the critical role of lactylation modifications in PDAC and offers potential therapeutic targets for cancer treatment. CUT&Tag technology efficiently and sensitively revealed the chromatin binding characteristics of H3K18la, showcasing its significant advantages in elucidating the mechanisms of histone post-translational modifications.
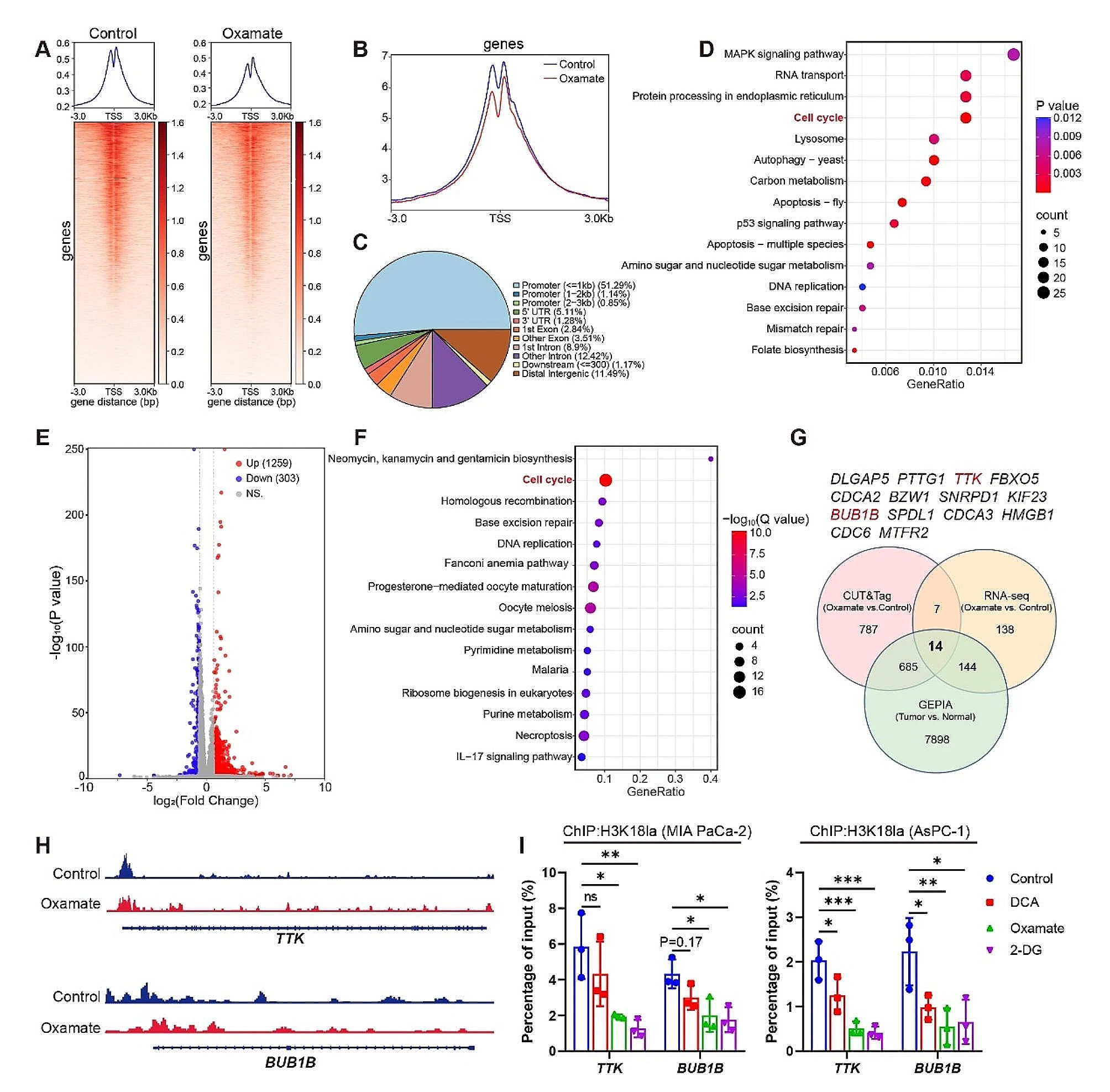
Li, F. et al. Mol Cancer. 2024.
CUT&Tag Analysis to Screen the Binding Sites of H3K18la
MtoZ Biolabs' CUT&Tag analysis service enables efficient exploration of the core mechanisms behind gene regulation. Our professional team and advanced platform deliver high-quality, reproducible research data to support breakthroughs in life sciences. Contact us to learn more about our CUT&Tag analysis service and how we can support your research needs.
How to order?







