Exosome Proteomics Service
Exosome proteomics is a comprehensive approach to studying the protein content of exosomes, which are small extracellular vesicles secreted by cells. This analysis utilizes advanced mass spectrometry and other proteomics technologies to identify and quantify proteins in exosomes, providing insights into their molecular composition and functional roles. By revealing the protein cargo of exosomes, exosome proteomics enables researchers to explore cell-to-cell communication, uncover disease-related pathways, and identify potential biomarkers.
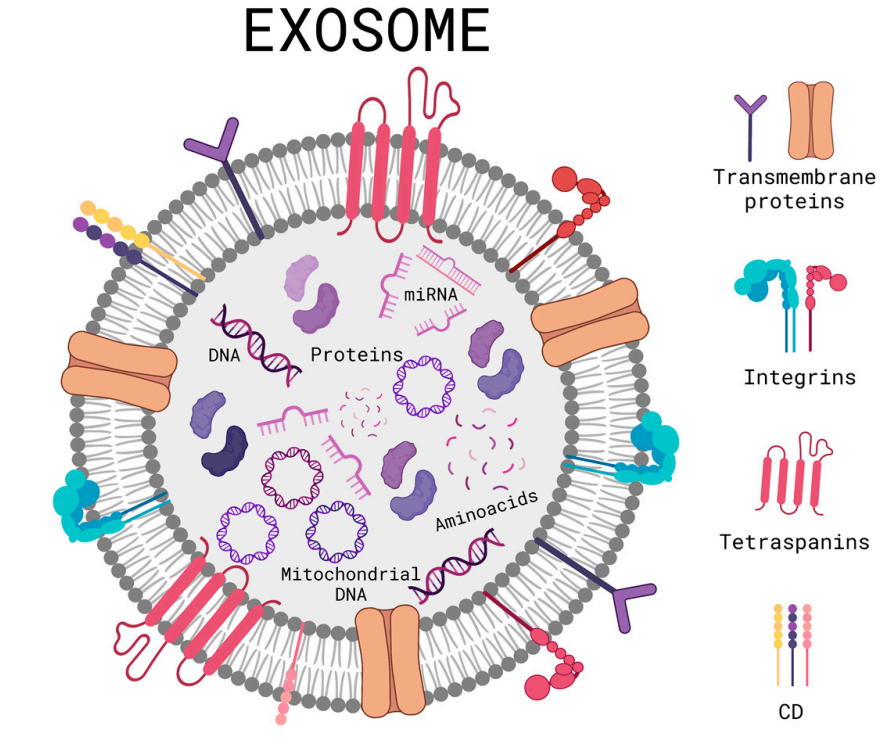
Tienda-Vázquez, MA. et al. Cells. 2023.
Figure 1. Schematic Diagram of a General Structure of an Exosome
The workflow of exosome proteomics analysis includes sample preparation, exosome enrichment, characterization, and proteomics analysis. Saliva samples undergo amylase removal, while serum samples are processed to remove albumin/IgG to eliminate interference. Exosomes are then enriched using specialized techniques and characterized through methods such as TEM observation and Western Blot validation. Finally, LC-MS/MS is used for high-sensitivity qualitative and quantitative analysis of exosomal proteins, revealing their composition and potential functions. Exosome proteomics service addresses key challenges in understanding the biological significance of exosomes and their roles in diseases through comprehensive analysis of the exosomal proteome. By identifying specific proteins associated with pathological processes, such as cancer metastasis or neurodegenerative disorders, it supports the discovery of diagnostic and therapeutic targets. Exosome proteomics service provides researchers with valuable insights into the molecular mechanisms driving disease progression and therapeutic responses, enabling a deeper understanding of exosome-mediated biology.
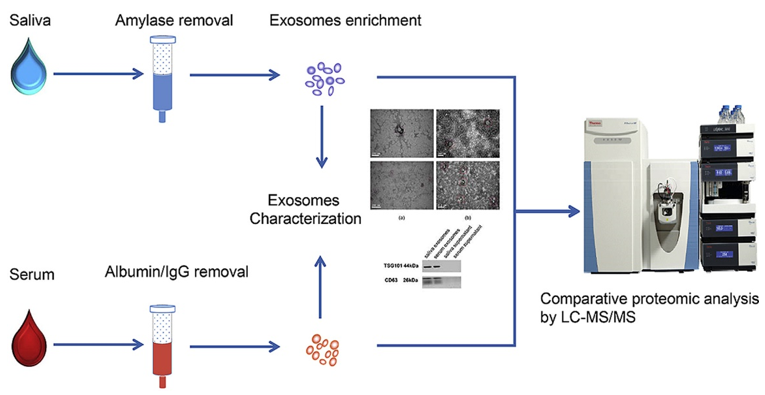
Sun, Y. et al. Anal. Chim. Acta. 2017.
Figure 2. Saliva and Serum Exosome Proteomics Workflow
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs offer advanced exosome proteomics service and have established an advanced proteomics platform, guaranteeing reliable, fast, and highly accurate analysis. Our experienced team specializes in mass spectrometry and proteomics, offering optimized workflows for high sensitivity and reproducibility in protein identification and quantification. With extensive expertise and a deep understanding of exosomal biology, MtoZ Biolabs offer reliable solutions to support research in disease mechanisms, biomarker discovery, and therapeutic development.
Service Advantages
1. Efficient Isolation and Purification Strategies
Our exosome proteomics service utilizes advanced isolation and purification techniques, including ultracentrifugation, density gradient separation, and immunoaffinity capture, ensuring high purity and integrity of exosomes in the samples, providing a reliable foundation for subsequent proteomics analysis.
2. In-Depth Proteome Coverage
Exosome proteomics service in MtoZ Biolabs combines high-resolution mass spectrometry with optimized protein extraction methods to comprehensively analyze key proteins within exosomes, helping researchers uncover the molecular composition and potential functions of exosomes in various diseases.
3. Accurate Quantification and Biomarker Discovery
Exosome proteomics service provided by MtoZ Biolabs employs advanced quantitative proteomics techniques, such as TMT labeling and label-free quantification, offering high sensitivity and accuracy for protein quantification, supporting the discovery of disease-associated biomarkers and their clinical translation.
Case Study
1. One-Pot Exosome Proteomics Enabled by a Photocleavable Surfactant
This study presents a one-pot exosome proteomics method utilizing a photocleavable surfactant, Azo, to streamline exosomal protein extraction, digestion, and analysis. The method simplifies sample preparation, enhancing throughput and reproducibility. Applied to mammary fibroblast-derived exosomes, the approach identified 3,466 proteins and quantified 2,288 proteins using reversed-phase liquid chromatography coupled with trapped ion mobility spectrometry (TIMS) quadrupole time-of-flight mass spectrometry (QTOF-MS). Notably, 91% of the identified proteins were annotated in ExoCarta and Vesiclepedia databases, including key exosomal markers CD63, PDCD6IP, and SDCBP. This method offers a fast, efficient, and reproducible strategy for comprehensive exosomal proteome profiling. Exosome proteomics service utilizes advanced mass spectrometry platforms to achieve high-throughput identification and quantification of exosomal proteins, providing reliable and in-depth analysis for applications in biomedical research, therapeutic development, and clinical diagnostics.
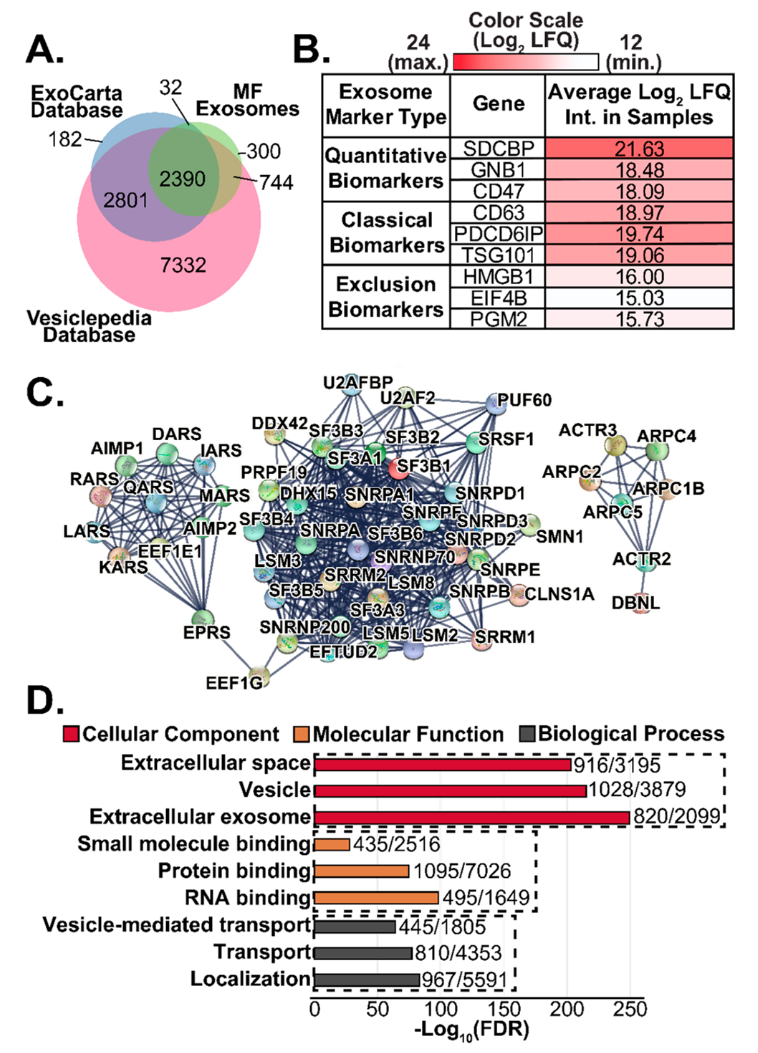
Buck, KM, et al. Anal Chem. 2022.
Figure 3. Exosome Proteomics Results Enabled by the Azo Surfactant
2. Comparative Proteomics Analysis of Exosomes Identifies Key Pathways and Protein Markers Related to Breast Cancer Metastasis
This study presents a comparative proteomics analysis of exosomes derived from mammary epithelial cells and breast cancer (BC) cell lines with varying metastatic potentials. A total of 2,135 unique proteins were confidently quantified from 20 exosome samples, including 94 of the TOP 100 markers listed in ExoCarta. Among them, 348 proteins showed significant alterations, with key metastasis-specific markers such as cathepsin W (CATW), magnesium transporter MRS2 (MRS2), syntenin-2 (SDCB2), reticulon-4 (RTN), and UV excision repair protein RAD23 homolog (RAD23B) identified. The expression levels of these proteins correlate with overall survival rates in clinical breast cancer patients, offering insights into the molecular mechanisms driving tumor progression and metastasis. Exosome proteomics service enables comprehensive analysis of exosomal protein profiles, allowing the identification of metastasis-specific biomarkers and altered signaling pathways through advanced mass spectrometry techniques, supporting targeted research into cancer progression and therapeutic development.
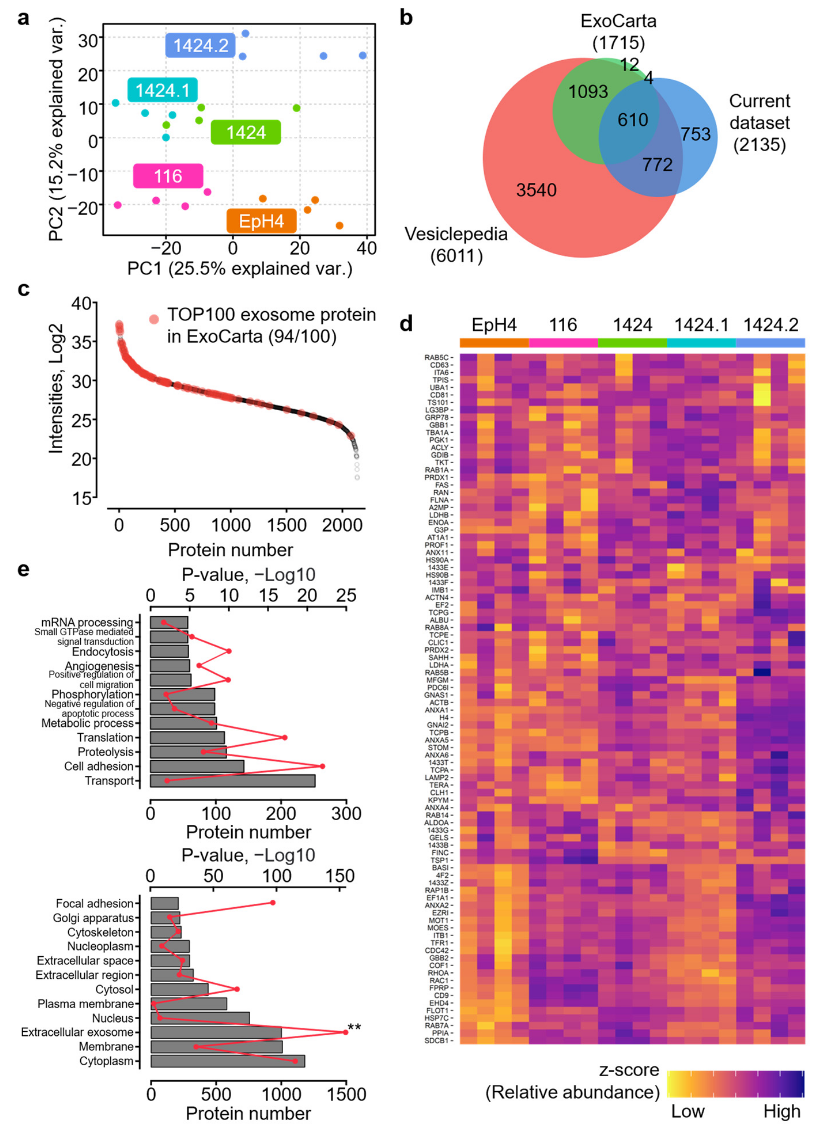
Shen, S. et al. Int J Mol Sci. 2023.
Figure 4. Proteomics Characterization of the Isolated Exosomes
Applications
1. Exosome Proteomics Service Can be Applied in the Identification of Tumor Biomarker
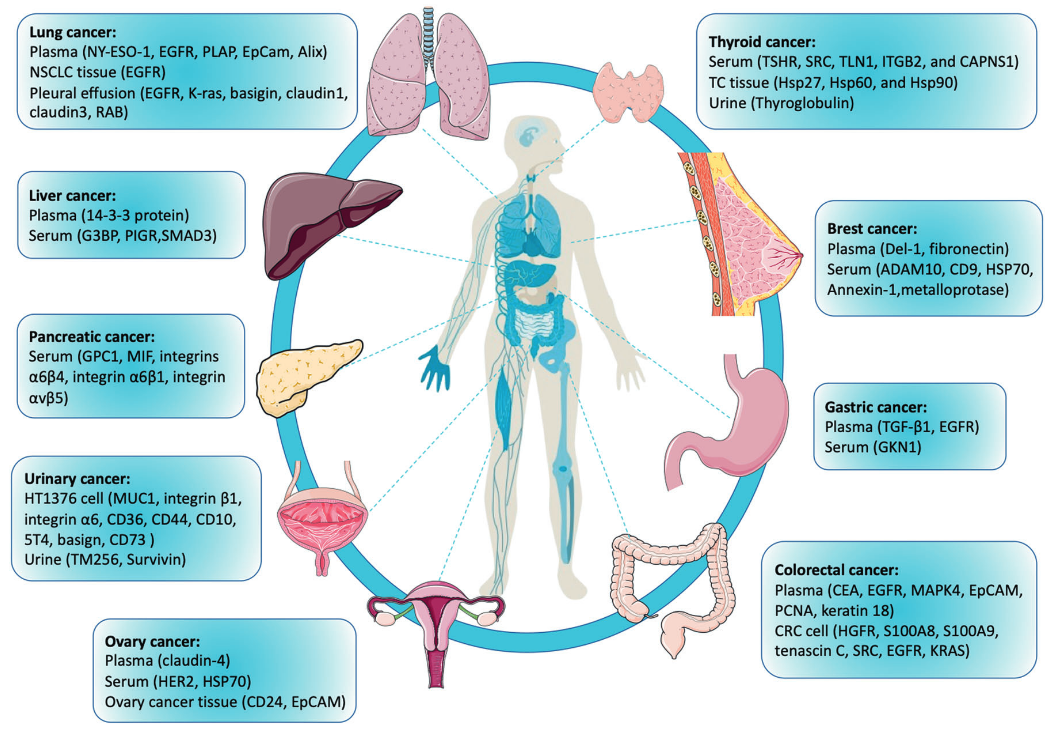
Wang, X. et al. Exp Mol Med. 2022.
Figure 5. Exosomal Protein Biomarkers Identified for Multiple Types of Human Cancer
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Liquid Chromatography and Mass Spectrometry Parameters
4. The Detailed Information of Exosome Proteomics
5. Mass Spectrometry Image
6. Raw Data
How to order?







