Functional Proteomics Analysis Service
Functional proteomics is a key branch of proteomics that aims to study the function, dynamic regulation and interaction network of proteins in biological systems. Functional proteomics analysis service emphasizes analyzing the biological roles of proteins, post-translational modifications (PTMs), protein-protein interactions (PPIs), and their dynamic changes in cells. By integrating high-precision mass spectrometry, protein interaction analysis techniques, and PTM analysis methods, functional proteomics analysis service provides comprehensive data support for uncovering the molecular mechanisms of life activities. Leveraging advanced chromatography and mass spectrometry platforms, the functional proteomics analysis service provided by MtoZ Biolabs can cover proteomics qualitative and quantitative analysis, protein interaction analysis, and PTM proteomics analysis, providing strong support for basic research and application development.
Gholap A D. et al. Springer, Singapore. 2023.
Analysis Workflow
1. Sample Preparation
Extract proteins from tissues, cells, or bodily fluids, removing interfering components to ensure reliability in downstream analysis.
2. Mass Spectrometry Analysis
Utilize high-resolution mass spectrometry for protein identification, quantification, and modification analysis.
3. Data Processing and Functional Annotation
Employ bioinformatics tools for data mining, annotating protein biological functions, associated pathways, and interaction networks.
4. Result Reporting
Deliver a comprehensive experimental report, including an identified protein list, post-translational modification sites, interaction networks, etc.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced functional proteomics analysis service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all functional proteomics analysis service data, providing clients with a comprehensive data report.
4. Comprehensive Functional Analysis: Reveal the biological roles of proteins from multiple dimensions, including expression levels, interactions, and post-translational modifications.
Sample Submission Suggestions
Sample Type
We accept a variety of sample types, including but not limited to cell lysate, tissue extracts, serum/plasma, etc.
Sample Quantity
It is recommended to provide ≥100 µg protein per group. The specific amount depends on the project. Please contact us for consultation before submitting samples.
Applications
Disease Research
Functional proteomics analysis service can reveal the molecular mechanisms in complex pathologies such as cancer, neurodegenerative diseases, and cardiovascular diseases.
Drug Development
Identify disease-associated key proteins, discover critical drug targets, and optimize drug screening processes.
Biomarker Discovery
Identify potential diagnostic or prognostic biomarkers for specific diseases through quantitative and functional studies.
Basic Scientific Research
Construct functional proteome networks of cells or tissues to understand the specific roles and regulatory mechanisms of proteins in life activities.
FAQ
Q. How to Apply Functional Proteomics Data to Disease Research?
Functional proteomics plays a vital role in disease research. By following steps, experimental data can be transformed into deeper insights into disease mechanisms and the discovery of therapeutic targets:
1. Data Analysis and Functional Annotation
Protein Function Annotation: Use Gene Ontology (GO) analysis to classify protein functions and determine their roles in biological processes such as metabolism and signal transduction.
Pathway Enrichment Analysis: Integrate KEGG or Reactome databases to analyze whether identified proteins are concentrated in specific signaling pathways or metabolic networks.
2. Key Protein Screening
Differential Expression Analysis: Compare healthy and diseased samples to identify significantly altered proteins, such as upregulated proteins in cancer.
Post-Translational Modifications (PTMs) Study: Investigate dynamic changes in PTMs under disease conditions to uncover regulatory mechanisms.
3. Disease-Related Network Construction
Protein-Protein Interaction (PPI) Network: Use tools like STRING or Cytoscape to construct PPI networks, identifying key nodes and core modules.
Functional Module Analysis: Focus on studying disease-related modules in the network to reveal their regulatory effects.
4. Target Validation and Biomarker Discovery
Target Validation: Verify the expression and function of key proteins through targeted mass spectrometry or Western blotting.
Biomarker Screening: Perform quantitative analysis to identify proteins with diagnostic or prognostic value, followed by validation using clinical samples.
5. Multi-Omics Integration
Integrate proteomics data with transcriptomics, metabolomics, and other omics to uncover multi-level molecular regulatory mechanisms, providing a panoramic view for disease research.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study used functional proteomics analysis technology to systematically identify the binding targets of various natural products such as eriocalyxin B and parthenolide with intracellular proteins, and analyzed the role of these targets in the anti-breast cancer signaling pathway. The results showed that these natural products affect key processes such as cell proliferation, apoptosis and metabolism through the modification and functional regulation of specific proteins.
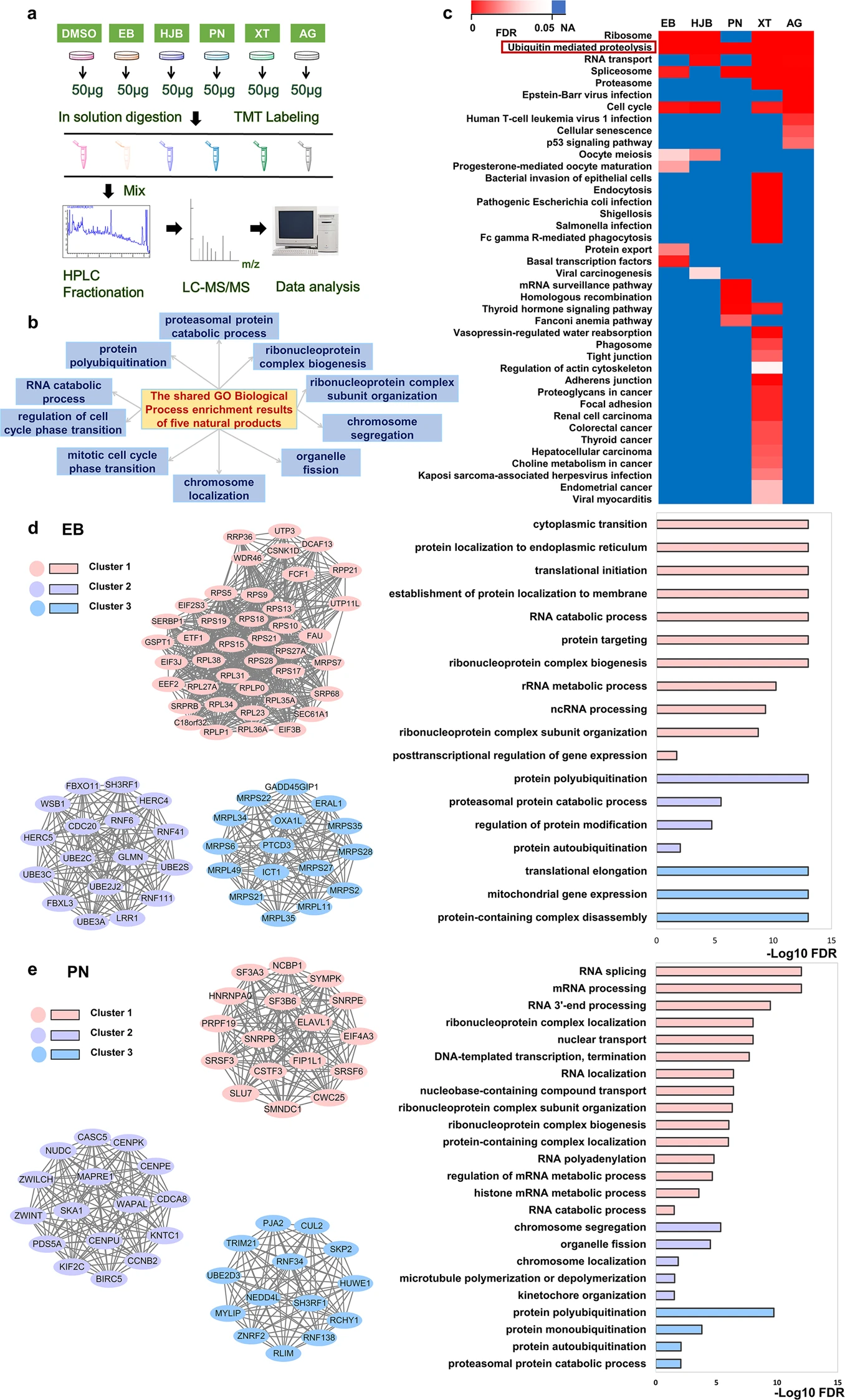
Zhao W S. et al. Acta Pharmacologica Sinica. 2023.
How to order?







