Introduction and Applications of CUT&Tag Technology
CUT&Tag (Cleavage Under Targets and Tagmentation) is an advanced technique designed for the study of DNA-protein interactions, primarily employed to identify the binding sites of transcription factors or histone modifications across the entire genome. The core principle of CUT&Tag involves the use of antibodies to specifically recognize target proteins or chromatin modifications, followed by the action of the Tn5 transposase, which generates sequencing-compatible tags at these binding sites. First introduced by Stephen Henikoff's group in 2019 as an enhancement and alternative to ChIP-Seq (Chromatin Immunoprecipitation Sequencing), CUT&Tag offers advantages including high sensitivity, low background noise, and reduced cellular input requirements. This technique has become an indispensable tool for investigating gene regulatory mechanisms, chromatin dynamics, and precision medicine. It finds applications in diverse fields, including epigenetics, transcription factor binding analysis, disease pathogenesis, drug target screening, and single-cell gene regulatory studies. MtoZ Biolabs offers comprehensive CUT&Tag analysis services by integrating a sophisticated Protein A/G-Tn5 fusion system with state-of-the-art high-throughput sequencing platforms. These services encompass sample preparation, library construction, sequencing, and bioinformatics analysis. With deep expertise in epigenomics, our technical team ensures precise and reliable analysis of protein-DNA interactions, supporting researchers in uncovering fundamental gene regulatory processes.
Background of CUT&Tag Technology
Histones compact and protect DNA by assembling nucleosomes. Post-translational histone modifications, such as acetylation and methylation, in coordination with DNA methylation, play crucial roles in regulating gene expression. These dynamic histone modifications are essential for maintaining cellular functions and are closely linked to various disease processes, including cancer. Understanding the nuanced interplay between histones and DNA is therefore of great scientific and clinical relevance for deciphering both normal cellular functions and disease mechanisms. Protein-DNA interactions are central to transcriptional regulation and are a prerequisite for initiating gene expression. Over time, scientists have developed several approaches to study these interactions, including gel shift assays, DNase I footprinting, methylation interference, in vivo footprinting, yeast two-hybrid screening, ChIP-Seq, and, more recently, CUT&Tag. This emerging technology is rapidly gaining prominence for its ability to precisely profile protein-DNA interactions and chromatin states with unparalleled resolution.
Comparison Between CUT&Tag and ChIP-Seq Technologies
ChIP-Seq combines chromatin immunoprecipitation (ChIP) with high-throughput sequencing to analyze protein-DNA interactions at a genome-wide scale. The process begins by enriching DNA fragments bound by the target protein through immunoprecipitation, followed by DNA purification and library preparation. These enriched DNA fragments are then subjected to high-throughput sequencing. By mapping the millions of obtained sequence reads to the genome, ChIP-seq enables the identification of genomic regions associated with histones, transcription factors, and other regulatory proteins. However, ChIP-seq has several limitations, including high cell input requirements, susceptibility to false positives and false negatives, difficulty in optimizing sonication parameters, heavy reliance on specific antibodies, low reproducibility, and a low signal-to-noise ratio.
CUT&Tag improves upon ChIP-seq by using a ProteinA/G-Tn5 fusion protein, which directly binds to the target protein without requiring crosslinking. This eliminates the need for formaldehyde crosslinking, which reduces background noise and enhances the resolution of binding site detection. The CUT&Tag protocol involves several key steps: First, ConA magnetic beads are used to capture glycoproteins on the cell membrane, and the cell nuclei are isolated or directly prepared from nuclear extracts, while digitonin is used to permeabilize the cell membrane, facilitating the accessibility of nuclear components. Next, a specific primary antibody (such as α-H3K27me3) is incubated with the sample, and non-specific binding is removed by washing. A secondary antibody is then added to ensure the precise binding of the antibody complex to the target region. The Protein A/G-Tn5 fusion protein then binds to the antibody complex, bringing the Tn5 transposase to the target DNA region. The reaction buffer, containing Mg²⁺, activates the Tn5 transposase, leading to DNA fragmentation and the insertion of sequencing adapters, completing the library preparation. Finally, high-throughput sequencing is performed using the Illumina platform, and subsequent bioinformatics analysis is conducted, including quality control, genome alignment, peak annotation, motif identification, GO analysis, and Kyoto Encyclopedia of KEGG pathway analysis. This comprehensive analysis allows for an in-depth exploration of transcription factor binding sites and the functional roles of chromatin modifications.
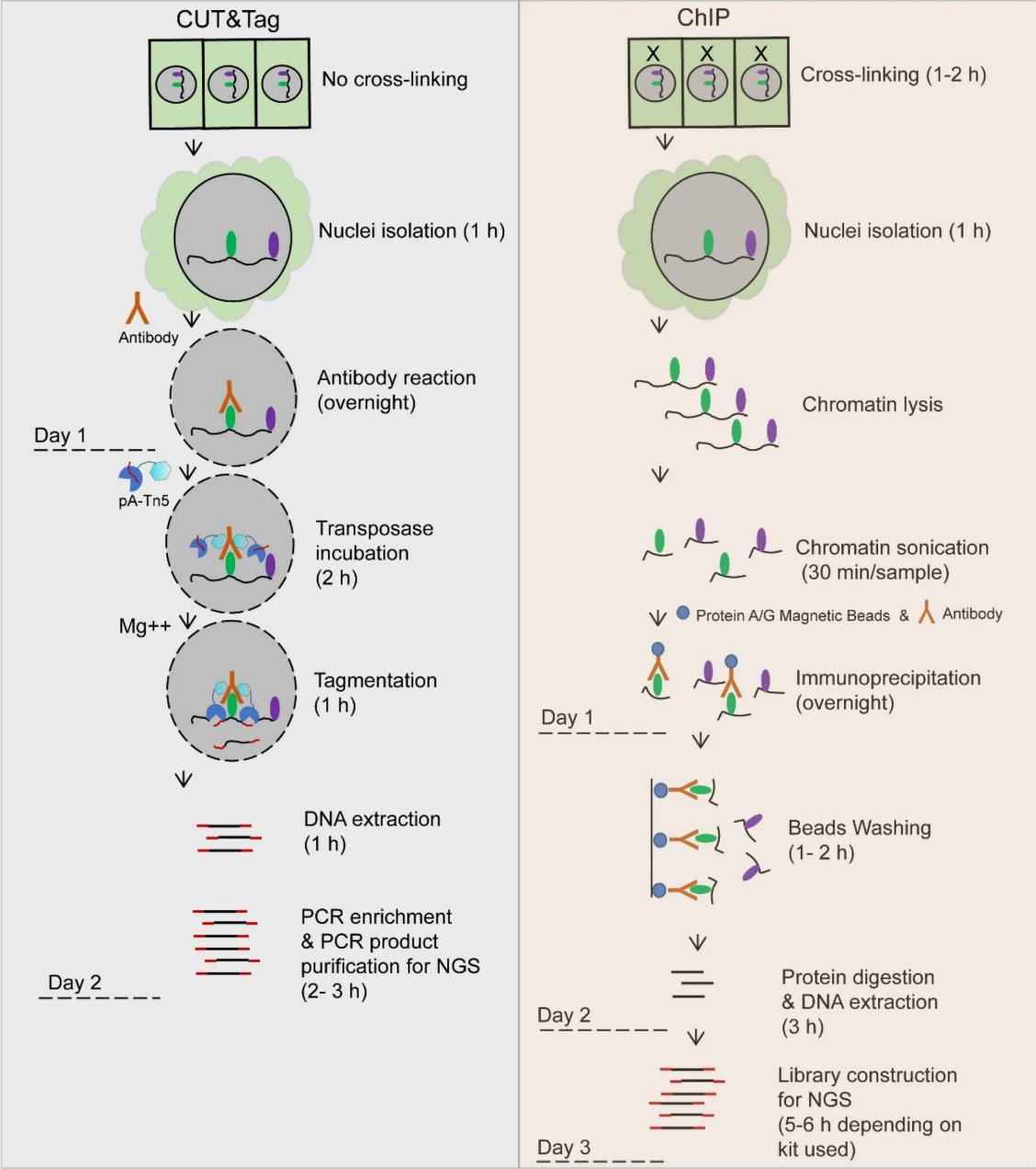
Figure 1. Comparative Overview of Technical Workflows (Plant Methods 16, 120 (2020))
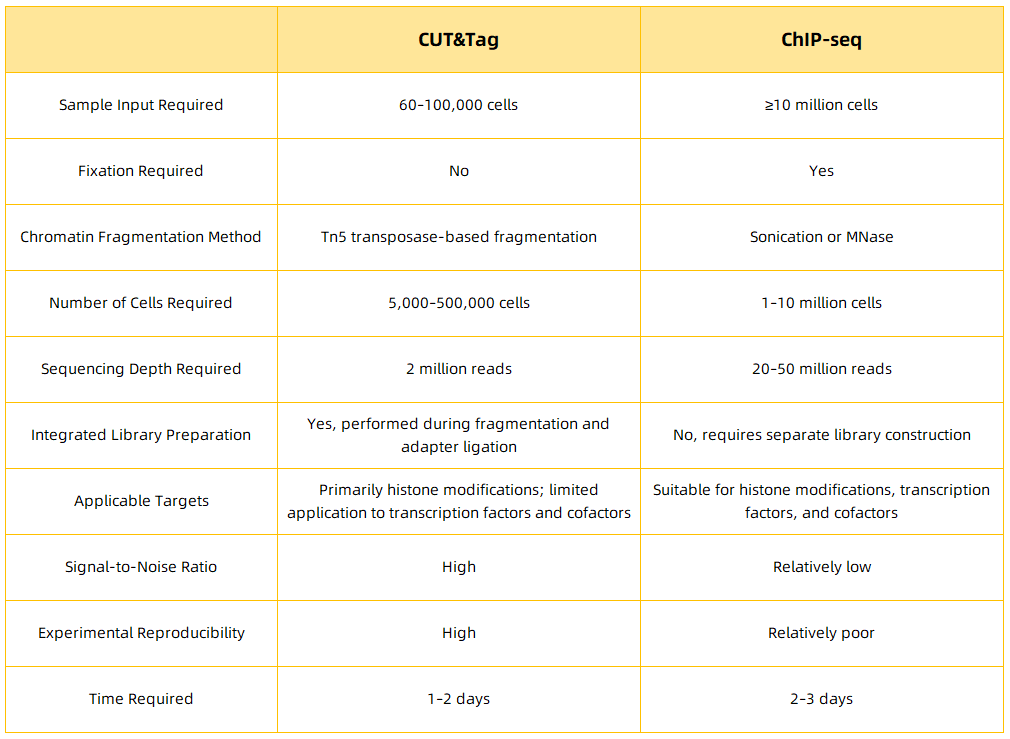
Figure 2.
Advantages of CUT&Tag Technology
1. Minimal Cell Input Requirement
CUT&Tag requires only a small number of cells, enabling analysis even at the single-cell level.
2. High Sensitivity and Low Background Noise
The method exhibits a high signal-to-noise ratio, ensuring reliable detection of protein-DNA interactions.
3. High Spatial Resolution
CUT&Tag precisely localizes transcription factor binding sites and histone modification regions on the genome.
4. Streamlined Workflow and Reduced Time Requirement
Compared to ChIP-Seq, CUT&Tag offers a simplified protocol with a significantly shorter experimental cycle.
Applications of CUT&Tag in Literature
CUT&Tag Reveals the Regulatory Mechanism of H3K18la in the Development and Progression of PDAC (Pancreatic Ductal Adenocarcinoma)
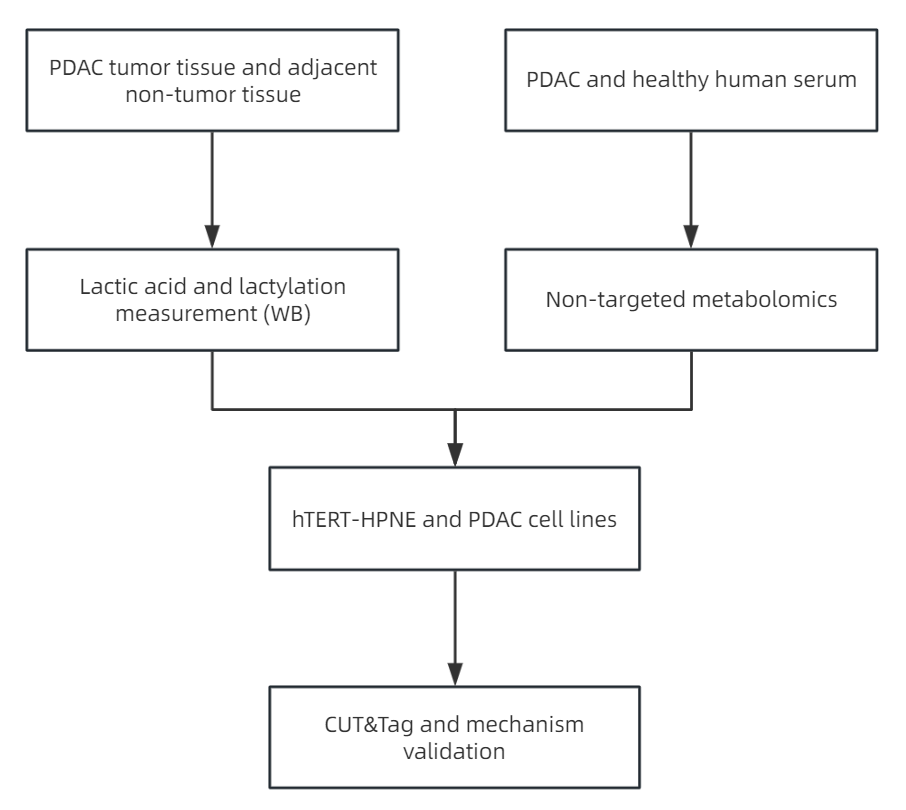
Figure 3.
Pancreatic cancer is an extremely invasive tumor of the digestive system and one of the most lethal malignancies in humans, with a notably low 5-year survival rate. Over 80% of pancreatic cancers are classified as pancreatic ductal adenocarcinoma (PDAC). Despite significant efforts to improve treatments in the past decade, the mortality rate for PDAC remains high. Therefore, a deeper understanding of the mechanisms driving PDAC growth and metastasis, as well as the identification of potential therapeutic targets and prognostic biomarkers, is urgently needed. PDAC cells utilize metabolic reprogramming to sustain malignant behaviors such as rapid proliferation, invasion, and other cellular processes. Enhanced glycolysis and lactate accumulation are hallmarks of various cancers. However, the precise role of metabolic reprogramming in reshaping epigenetic modifications, particularly lactylation in PDAC, remains poorly understood. In this context, the study titled "Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma," published in Molecular Cancer (2024, IF: 27.7), presents the following key findings:
1. Elevated Histone Lactylation Levels Correlate with Poor Prognosis in PDAC Patients
(1) In pancreatic tissues from PDAC patients, lactate levels were significantly higher than in adjacent normal tissues (Figure 4A). Similarly, PDAC cell lines such as MIA PaCa-2, PANC-1, AsPC-1, and PL45 exhibited higher lactate production compared to the human pancreatic ductal epithelial cell line hTERT-HPNE (Figure 4B). Untargeted metabolomics analysis of serum samples from both PDAC patients and healthy controls revealed significant metabolic changes. A total of 222 metabolites showed significant differences between the tumor and healthy groups, with 100 metabolites lower and 122 metabolites higher in the tumor group (Figure 4C). KEGG analysis of these metabolites indicated an enrichment of central carbon metabolism (Figure 4D). Notably, lactate was one of the metabolites identified.
(2) Given the substantial production of lactate as a substrate for histone lactylation, this study examined protein lactylation levels in 5 pairs of PDAC and adjacent non-cancerous tissues. Higher levels of global pan-lysine lactylation (Pan Kla) were observed in PDAC tissues (Figure 4E). Histone modifications, which play a fundamental role in most biological processes, are important in regulating tumorigenesis. H3K18la, a specific histone modification, was chosen for further investigation. Consistent with earlier findings, H3K18la levels were significantly elevated in PDAC tissues (Figure 4E). Additionally, Pan Kla and H3K18la expressions were significantly upregulated in all 4 PDAC cell lines compared to normal pancreatic ductal epithelial cell lines (Figure 4F).

Figure 4.
(3) To further investigate the clinical significance of H3K18la, this study performed immunohistochemical staining on 74 PDAC and 72 adjacent non-cancerous tissue samples (excluding shedding sections, non-pancreatic ductal tissues, and morphologically abnormal adjacent tissues). As shown in Figures 5G and 5H, H3K18la levels were significantly higher in PDAC tissues compared to normal tissues. Among PDAC patients, 51.4% of samples exhibited high levels of H3K18la, which was significantly more common than in the normal group (Figure 5I). The increased levels of H3K18la were positively correlated with advanced AJCC stages (Figure 5J). To further evaluate the prognostic value of H3K18la, Kaplan-Meier survival curves were constructed based on H3K18la levels. Log-rank tests indicated that elevated H3K18la levels may be associated with poor prognosis (Figure 5K).

Figure 5.
2. Glycolysis Inhibition Reduces Histone Lactylation and Suppresses PDAC Cell Proliferation and Migration
(1) To investigate the impact of glycolysis on histone lactylation, this study utilized different glycolysis inhibitors-DCA, Oxamate, and 2-DG-to knock down LDHA (the enzyme responsible for converting pyruvate to lactate) in MIA PaCa-2 and AsPC-1 cells. As the concentration of the inhibitors increased, the levels of pan-lysine lactylation (Pan Kla) and H3K18la progressively decreased. Additionally, si-LDHA effectively reduced histone lactylation, whereas treatment with Nala (lactate salt) significantly elevated histone lactylation levels.
(2) The biological effects of histone lactylation on cell proliferation were further explored. Treatment with DCA, Oxamate, or 2-DG led to a marked reduction in both cell viability and colony formation ability. Furthermore, silencing of LDHA significantly inhibited the growth and colony formation of both MIA PaCa-2 and AsPC-1 cells, while Nala treatment alleviated the inhibitory effects of LDHA silencing on cell proliferation.
(3) These results suggest that histone lactylation plays a critical role in the initiation and progression of PDAC, and that inhibiting histone lactylation may offer potential anti-tumor activity in PDAC.
3. H3K18 Lactylation Activates the Transcription of TTK and BUB1B in PDAC
(1) To explore how H3K18 lactylation (H3K18la) affects PDAC progression, this study performed CUT&Tag analysis using an anti-H3K18la antibody. In the treated group, a significant reduction in the transcription start site (TSS) region was observed (Figures 6A and 6B), with approximately 53% enrichment in the promoter region (Figure 6C). KEGG pathway analysis of the downregulated peaks in specific promoter regions of the treated group revealed enrichment in RNA transport, cell cycle, and cancer-related pathways, such as the MAPK signaling pathway (Figure 6D).
(2) Next, this study identified potential target genes regulated by lactylation through RNA sequencing (RNA-seq). Compared to the control group, Oxamate treatment upregulated 1259 genes and downregulated 303 genes in PDAC cells (Figure 6E). KEGG analysis of the downregulated genes also revealed enrichment in cell cycle pathways in the Oxamate-treated group (Figure 6F). Gene Ontology (GO) analysis of both the CUT&Tag and RNA-seq data further confirmed enrichment in cell cycle-related processes.
(3) By combining CUT&Tag data, RNA-seq results, and the GEPIA database, this study identified 14 genes that were highly expressed in PDAC and regulated by H3K18la, with a strong association to cell cycle progression. ChIP-qPCR validation demonstrated that reduced levels of H3K18la at the TTK and BUB1B promoter regions were associated with the use of glycolysis inhibitors. Moreover, these inhibitors downregulated both the mRNA and protein levels of TTK and BUB1B, indicating that H3K18la regulates the expression of these genes via the glycolytic pathway, thereby influencing PDAC progression.
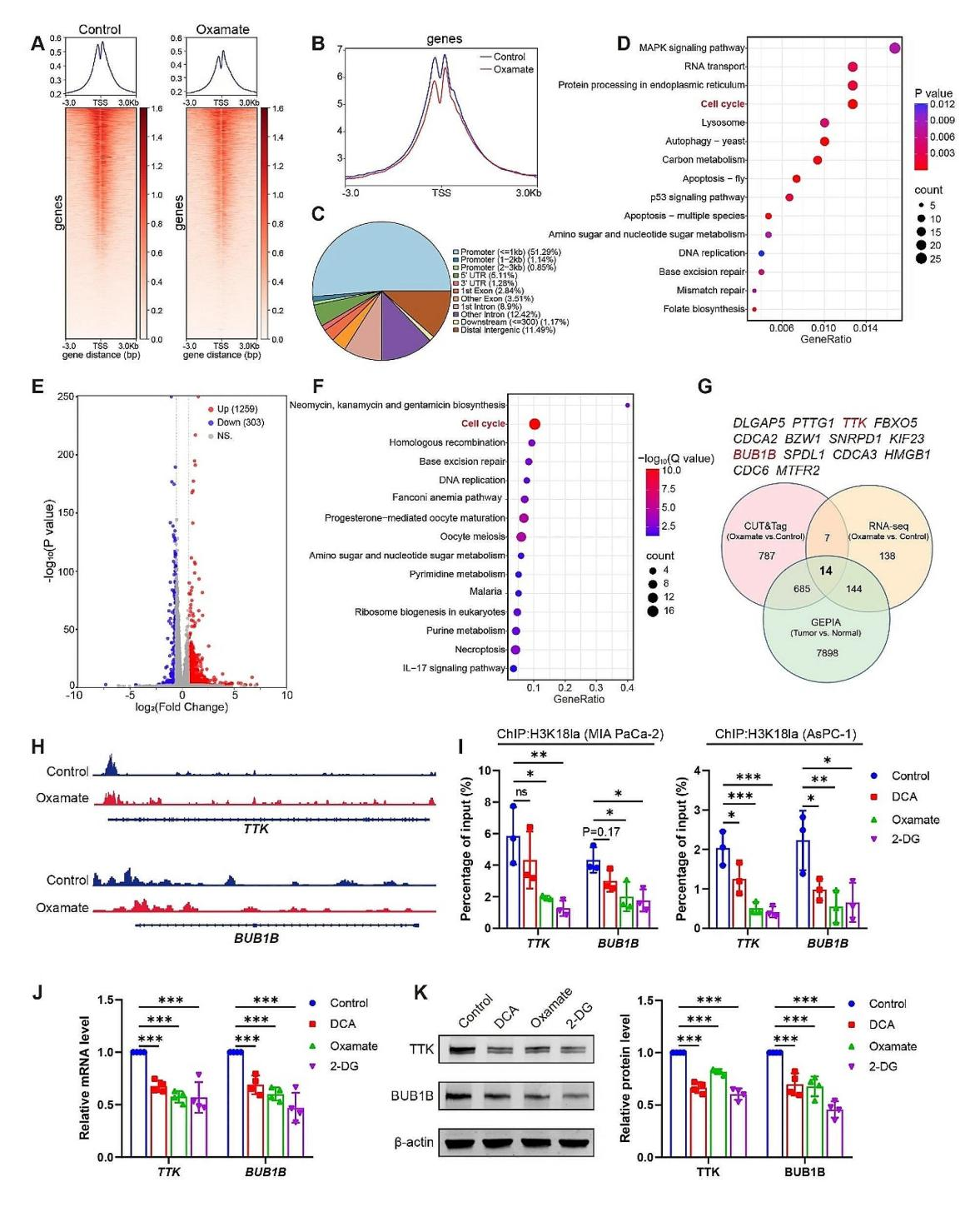
Figure 6.
Studies have demonstrated that CUT&Tag is extensively utilized to map the distribution of specific chromatin modifications, such as H3K27me3 and H3K4me1, across the genome. Through CUT&Tag, the precise localization of various chromatin modification marks can be clearly identified, contributing to a deeper understanding of the epigenetic mechanisms governing gene regulation. Moreover, CUT&Tag is also essential in other research fields. In the identification of transcription factor binding sites, CUT&Tag enables the accurate mapping of transcription factor binding across the genome, shedding light on their role in the regulation of gene expression. In the context of single-cell epigenomics, the low sample requirements of CUT&Tag allow single-cell analysis to reveal cellular heterogeneity and differences in chromatin states within a population of cells.
CUT&Tag technology is rapidly driving advancements in the field of epigenetics. MtoZ Biolabs offers professional and comprehensive CUT&Tag analysis services that empower you to accurately characterize chromatin states, uncover the underlying mechanisms of gene regulation, and provide robust support for research into disease mechanisms and drug development. Our services encompass the entire workflow, from sample preparation and antibody optimization to high-throughput sequencing and data analysis, ensuring the delivery of high-quality data on chromatin modifications and transcription factor binding sites. Partner with MtoZ Biolabs to elevate your research to new heights!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







