Label Free Quantification Proteomics Service
- Cell Samples: Lysates with a protein concentration of 1-5 mg/mL, minimum volume 100 µL. Fresh or snap-frozen samples are required to avoid degradation.
- Tissue Samples: Fresh or snap-frozen tissues with a protein concentration of 1-5 mg/mL.
- Biological Fluids: Examples include serum, plasma, and urine; ≥1 mL of fresh or frozen material is recommended.
- Purified Proteins: Protein concentration ≥1 mg/mL, purity ≥90%, free of aggregation or degradation.
- For custom requirements, please contact our technical team.
Label Free quantification proteomics is a cutting-edge mass spectrometry-based protein quantification method that eliminates the need for isotopic or chemical labeling. By directly analyzing endogenous peptide signals, Label Free quantification proteomics accurately quantifies protein expression levels across various conditions. This technique leverages high-resolution mass spectrometry for peptide detection and identification, followed by quantitative analysis using database searches. Comparing peptide signal intensities across samples enables precise determination of relative protein abundance.
Characterized by high throughput, broad proteomic coverage, and applicability to complex samples, this method has become indispensable in biological research, disease mechanism studies, and drug development. MtoZ Biolabs, with its advanced mass spectrometry platforms and expert technical team, offers highly sensitive and reproducible Label Free quantification proteomics service. From sample preparation to data interpretation, we deliver comprehensive solutions to accelerate your research.
Why Choose MtoZ Biolabs?
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Label Free quantification proteomics platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrate all Label Free quantification proteomics data providing clients with a comprehensive data report.
4. Multi-Sample Compatibility: Our platform supports a wide variety of biological samples, including cells, tissues, blood, and bodily fluids, providing flexibility to accommodate diverse research demands.
5. Customized Service: Tailored to the specific research needs of our clients, we offer flexible experimental design and personalized data analysis to ensure the achievement of research goals to the fullest extent.
Sample Submission Suggestions
To ensure optimal results and accurate protein quantification, proper sample handling and submission are essential. Following these guidelines will help maintain the integrity of your samples and ensure high-quality data from our Label Free quantification proteomics service. Please review the recommendations below for each sample type:
Applications
MtoZ Biolabs' Label Free quantification proteomics service is widely utilized across various research areas, including disease mechanism exploration, biomarker discovery, drug development and target validation, systems biology, and agricultural and environmental sciences. This versatile approach provides comprehensive insights into protein expression and function, enabling breakthroughs and advancing research across these key fields.
Case Study
Case 1
Using Label Free quantification proteomics, this study compared four strains of Lactococcus lactis (NCDO2118, IL1403, NZ9000, MG1363), identifying 586 core proteins associated with key functions such as stress resistance and probiotic properties. KEGG pathway analysis highlighted significant pathways, including ribosomal function, central metabolic pathways, and pyruvate metabolism, shedding light on the metabolic adaptations of the strains. Additionally, strain-specific proteins and unannotated proteins were identified, which may contribute to unique probiotic functions. These findings deepen our understanding of Lactococcus lactis biology and provide a foundation for optimizing its biotechnological applications, particularly in the development of functional foods and probiotics.
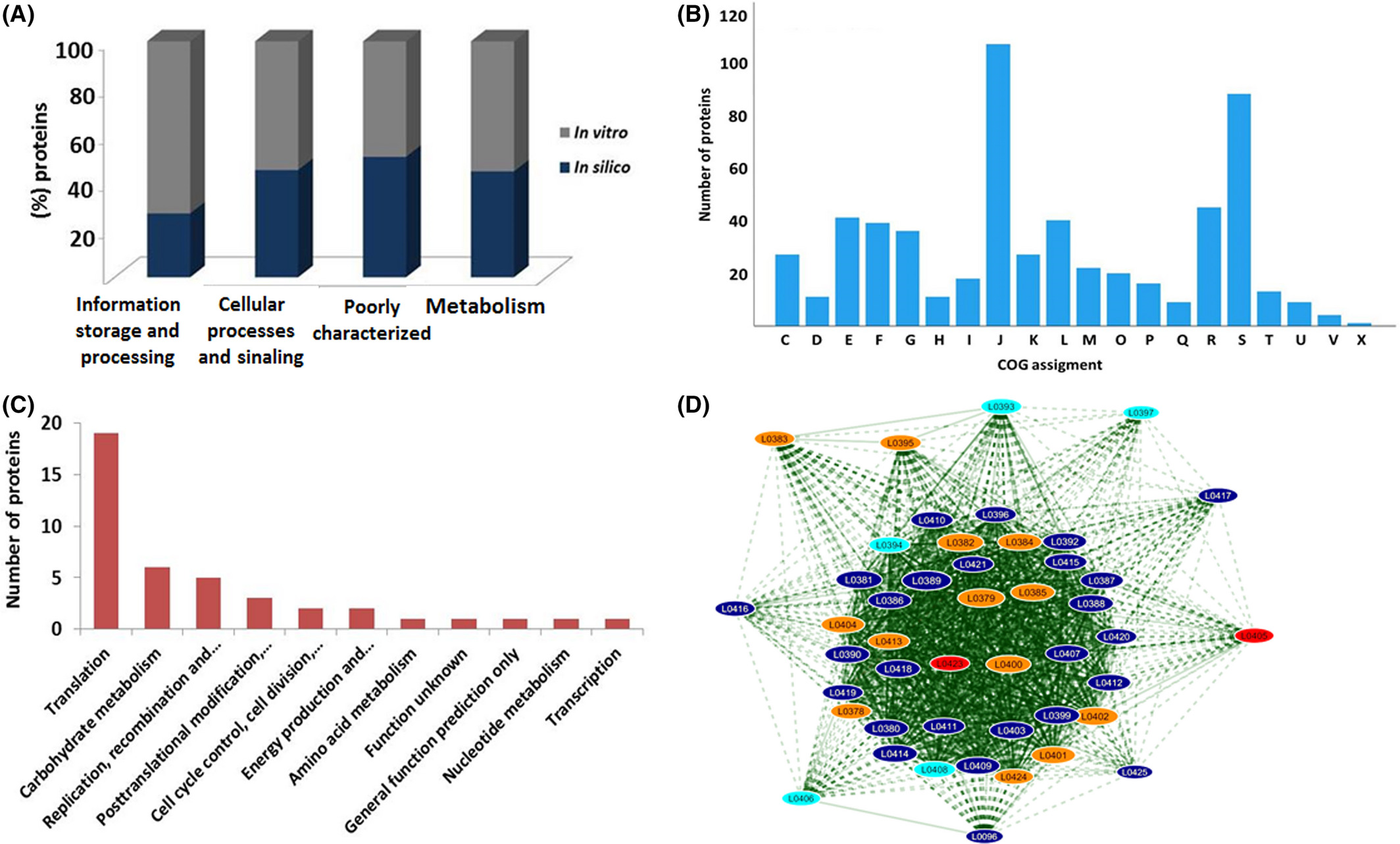
Silva, W. M. et al. Microb. Biotechnol. 2018.
Case 2
This study used Label Free quantification proteomics combined with bioinformatics to explore protein expression changes in chronic and binge alcohol-induced liver disease mouse models. The analysis identified 87 upregulated and 133 downregulated differentially expressed proteins (DEPs). Functional annotation revealed that these proteins are primarily involved in critical biological processes such as protein binding, metabolism, signal transduction, and immune response. Key proteins, including TRIP12, NDUFAF3, and GMPS, were validated via qPCR, confirming their altered expression patterns. These results provide significant insights into the molecular mechanisms of alcoholic liver disease and offer potential targets for developing novel diagnostic markers and therapeutic strategies.
Zhang, Y. et al. Mol. Med. Rep. 2018.
Case 3
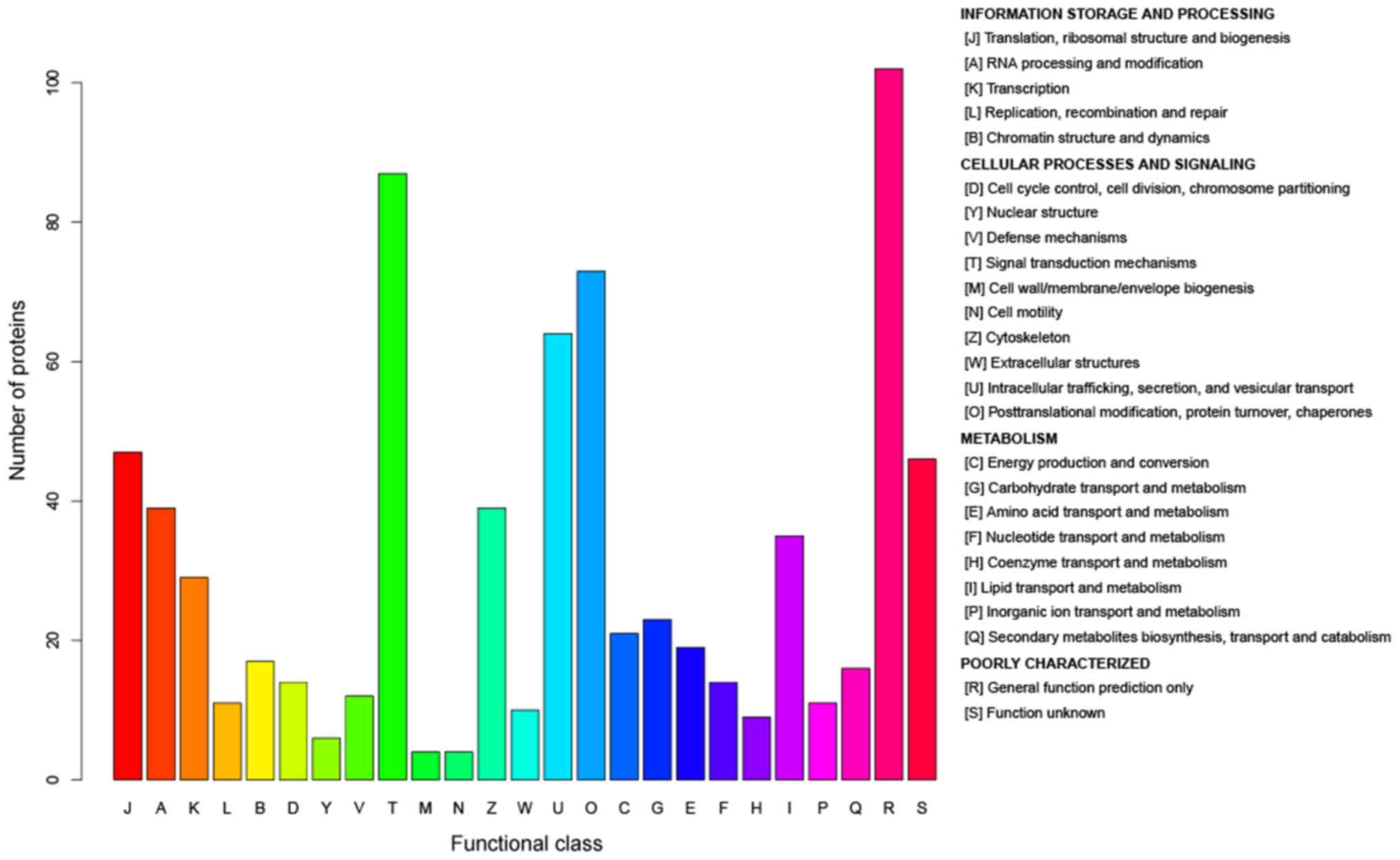
This study employed Label Free quantification proteomics to analyze Longissimus muscle and plasma samples from 20 Charolais × Aubrac crossbred heifers. A total of 268 proteins in muscle and 136 proteins in plasma were identified, among which 71 muscle proteins and 21 plasma proteins exhibited significant differences correlated with tenderness and toughness traits. Key proteins such as ACTN2, TNNC1, OGDH, HPX, ADSSL1, OGN, GOT1, and VCL in muscle, as well as HPX and OGN in plasma, were identified as markers strongly associated with tenderness. Functional analyses revealed that these proteins are involved in essential biological processes, including muscle structure maintenance, metabolic regulation, and oxidative stress response. This study provides valuable insights into the molecular mechanisms underlying beef tenderness and identifies potential biomarkers for improving meat quality.
Boudon, S. et al. J Proteomics. 2020.
Case 4
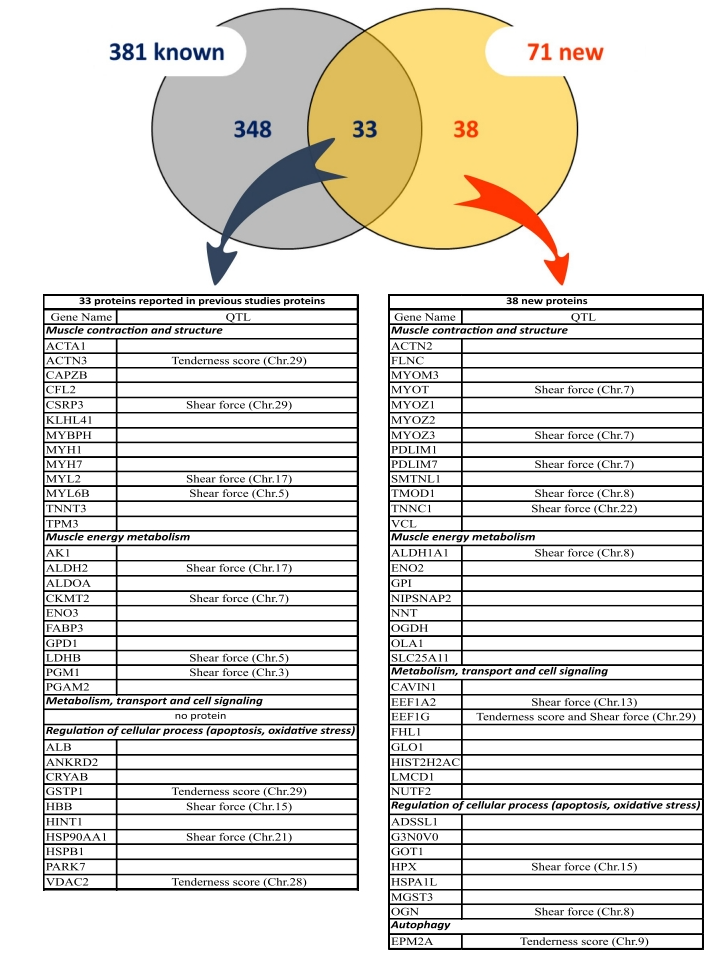
This study examined the molecular responses of maize (Zea mays) roots during early symbiosis with the endophytic bacterium Herbaspirillum seropedicae using Label Free quantification proteomics. A total of 123 differentially expressed proteins were identified, including 34 enzymes upregulated during the early stages of bacterial colonization. These enzymes are primarily involved in critical pathways such as carbohydrate metabolism, amino acid metabolism, and stress response mechanisms. The findings illuminate the molecular regulatory networks in maize roots during symbiosis, providing new perspectives on plant-microbe interactions and actionable insights for enhancing crop productivity and stress resistance strategies.
Nunes, R. de O. Proteomics. 2021.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
How to order?







