MS-Based Protein N-Terminal Sequence Analysis Service
Mass spectrometry N-terminal sequence analysis is a method that utilizes mass spectrometry technology to efficiently and accurately determine the amino acid sequence of the N-terminus of proteins/peptides. Due to its high sensitivity, high resolution, and high throughput, it can be used to analyze complex protein/peptide mixtures, as well as identify modifications, mutations, and other sequence variations. Compared to traditional Edman degradation, mass spectrometry offers higher throughput and sensitivity, enabling the analysis of more complex samples and larger proteins.
Based on high-resolution mass spectrometry for N-terminal sequencing, the sample is processed with SDS-PAGE to separate the target protein followed by enzyme digestion. The digested products are analyzed using a Q-Exactive series mass spectrometer (Thermo Fisher Scientific), with peptide fragments in the mass spectrum breaking down into b and y ions' secondary mass spectra. By comparing these spectra with the theoretical amino acid sequence of the protein, the N-terminal position of the protein is determined. MtoZ Biolabs combines high-performance liquid chromatography and high-resolution mass spectrometry, based on the principle of N-terminal sequencing, to establish a mass spectrometry-based N-terminal sequence analysis platform, providing you with high-quality mass spectrometry N-terminal sequence analysis services. We use two different proteolytic cleavage methods to produce two different lengths of N-terminal peptide segments, which are mutually verified to finally determine the protein N-terminal sequence. Welcome your consult for more detailed information!
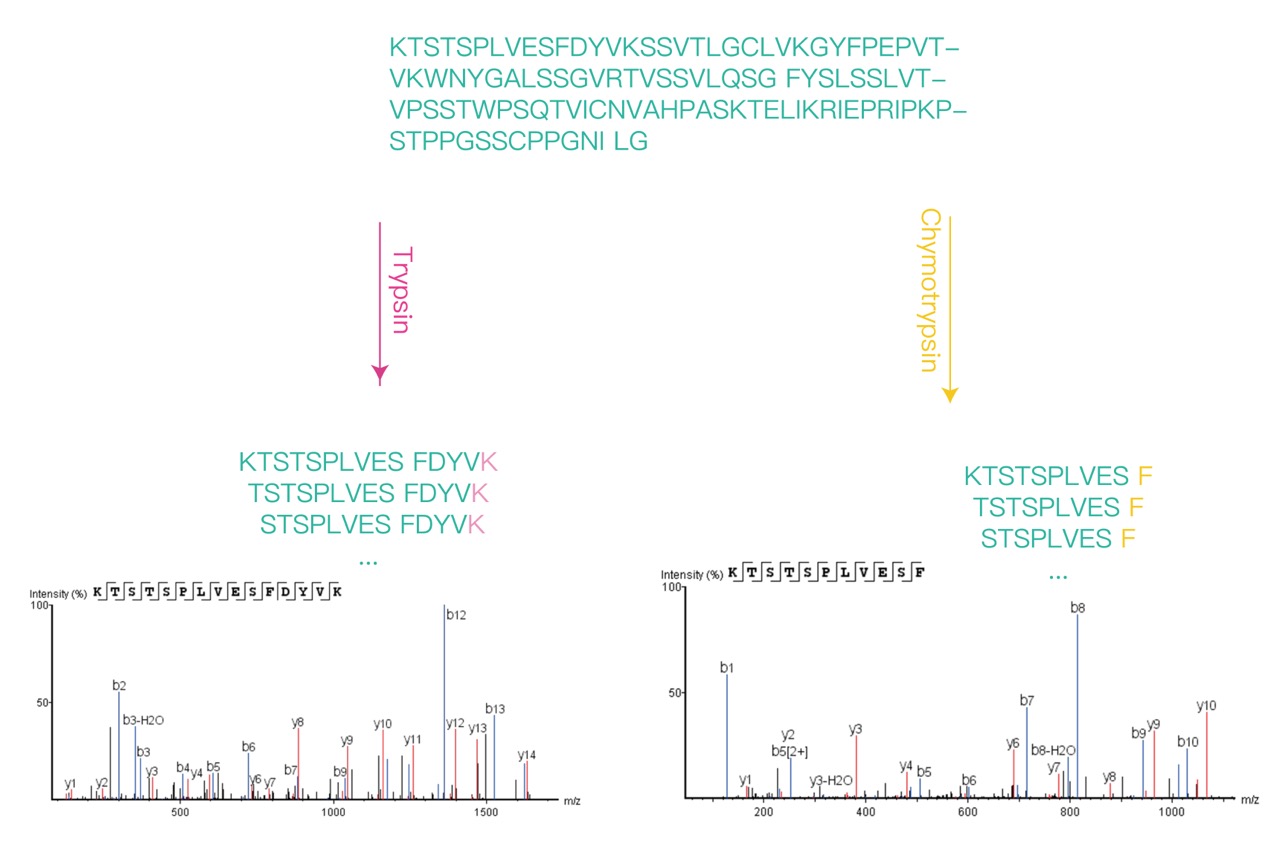
Figure 1. Comparison of Two Enzymatic Cleavage Methods in Mass Spectrometry N-terminal Sequence Analysis
Experimental Instruments
1. High-performance Liquid Chromatography Tandem Mass Spectrometer: High-performance Liquid Chromatograph-Easy-nLC 1200
2. Electrospray-hybrid Ion Trap Orbitrap Mass Spectrometer - Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer
Service Advantages
1. High-resolution Mass Spectrometry-Based N-terminal Sequencing for Analysis of N-terminal Capping and Post-translational Modifications (PTMs), Determining the Protein N-terminal Starting Point
2. Support for N-terminal Dimethylation Labeling in Complex Proteins by our Company, Involving Analysis of Labeling Positions to Determine the N-terminal Starting Points in Complex Protein Samples
Case Study
N-terminal Amino Acid Sequence Analysis of a Protein Drug Sample:
After trypsin digestion, the sample is analyzed by LC-MS/MS, and the mass spectrometry data is matched with a theoretical database, obtaining an MS2 spectrum consistent with the theoretical N-terminal sequence (HYAHVDCPGHADYVK), as shown below:
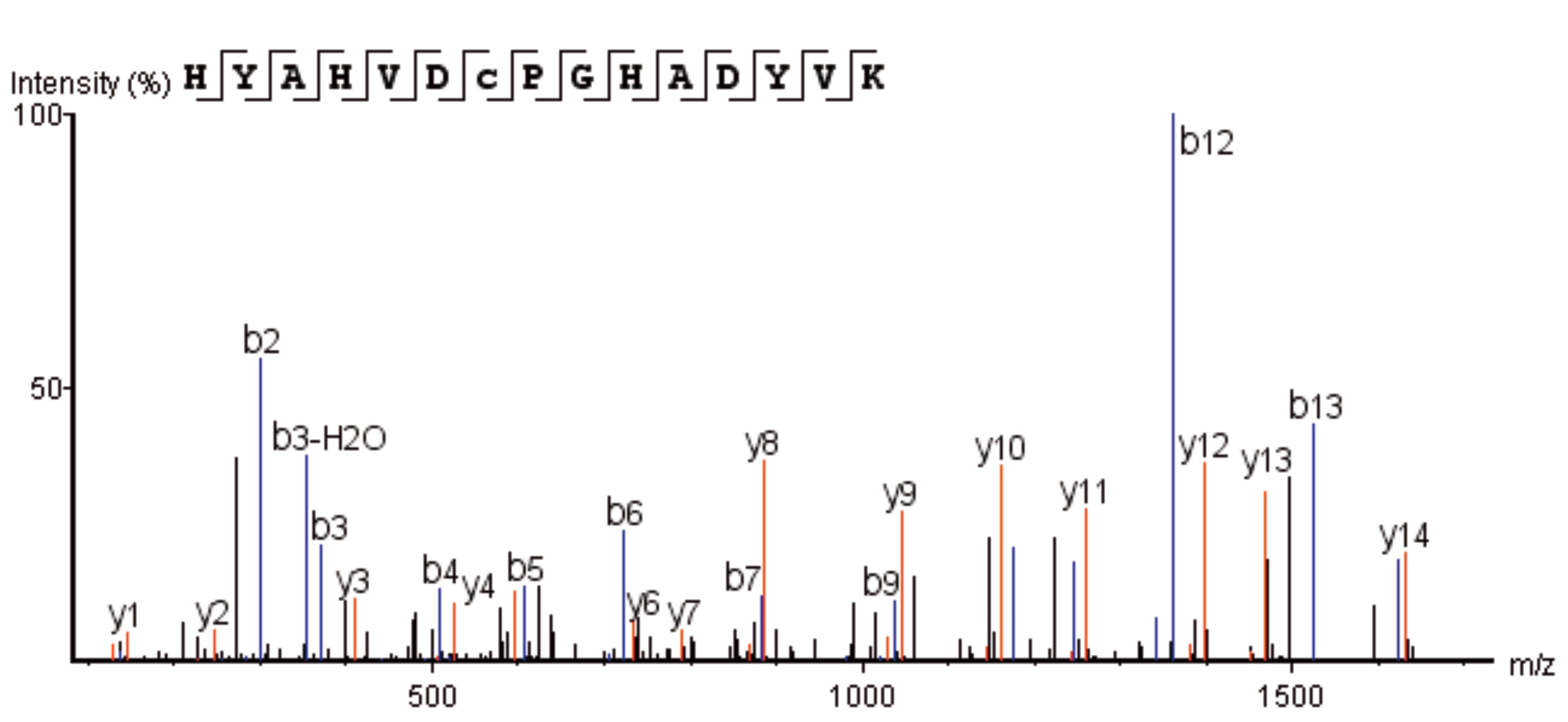
Figure 2. Secondary Mass Spectrum of N-terminal Sequence
From the analysis above, it can be concluded that the N-terminal starting amino acid of this protein drug is histidine, with the N-terminal sequence as HYAHVDCPGHADYVK.
Services at MtoZ Biolabs
1. Experimental Procedures
2. Mass Spectrometric Parameters
3. Detailed Information on Mass Spectrometry N-terminal Sequence Analysis
4. Mass Spectrometric Images
5. Raw Data
How to order?







