Mass Spectrometry-Based Peptide Identification Service
Peptides, structurally akin to proteins, are molecules composed of amino acid sequences. They play pivotal roles in regulating blood pressure, modulating pain perception, and managing glucose metabolism, yet are significantly smaller than proteins. Endogenous peptides typically arise from the cleavage of precursor proteins by various enzymes. Research into the collective peptide expression under specific physiological conditions is termed peptidomics.
Peptide identification aims to achieve "Peptide-Spectrum Matches" by analyzing peptide sequence information to ascertain sequences or post-translational modifications. Tandem mass spectrometry, extensively employed in peptide identification, combines with bioinformatics for peptide sequencing in proteomics. This involves matching experimental spectral data against theoretical data from protein databases. A current focus of research is matching proteins with unknown sequences to database entries.
Analysis Workflow
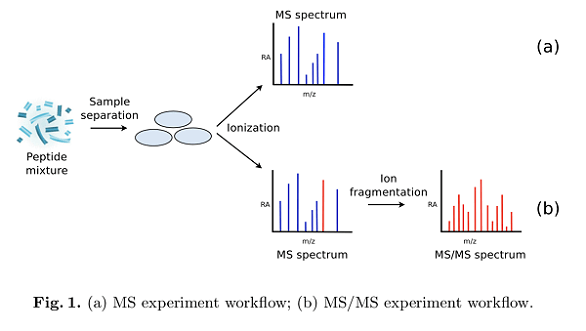
Figure 1. Peptide Identification Process
Services at MtoZ Biolabs
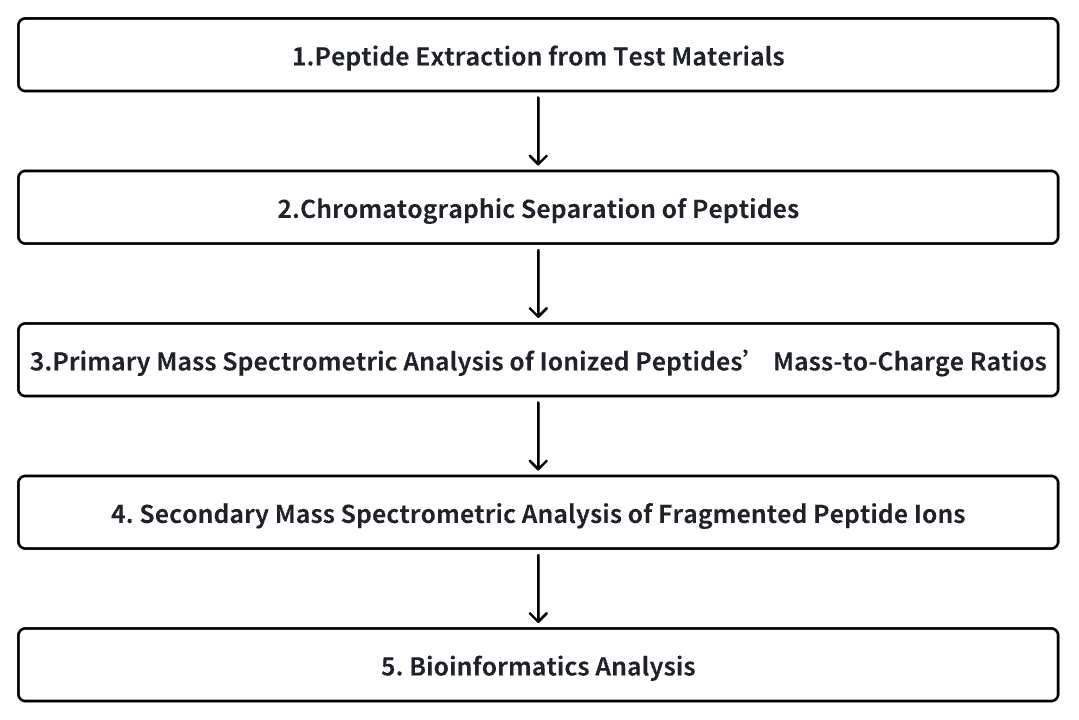
1. Peptide Extraction from Test Materials
2. Chromatographic Separation of Peptides
3. Primary Mass Spectrometric Analysis of Ionized Peptides' Mass-to-Charge Ratios
4. Secondary Mass Spectrometric Analysis of Fragmented Peptide Ions
5. Bioinformatics Analysis
Sample Submission Requirements
Gel sealing in plastic film or gel strips in sealed tubes. Maintain moisture in sealed samples without excessive water or buffer.
Considerations
1. Mandatory Glove Use for Gel Cutting
2. Cleanliness of Cutting Blades
3. Precision in Cutting Gel Strips to Minimize Background Noise
4. Stringent Avoidance of Keratin Contamination from Handling and Environmental Exposure
Deliverables
In the technical report, MtoZ Biolabs will provide you with detailed technical information, including:
1. Experimental Procedures
2. Mass Spectrometry Parameters
3. Mass Spectrometry Images
4. Raw Data
5. Detailed Information of Identified Proteins
How to order?







