Protein-Protein Interaction Service
- Stable vs. Transient: Stable protein-protein interactions are typically involved in forming stable protein complexes, while transient protein-protein interactions are often involved in dynamic regulatory processes, such as those in signaling pathways.
- Direct vs. Indirect: Direct protein-protein interactions occur through physical interactions between proteins, while indirect protein-protein interactions involve intermediary molecules.
- In vivo vs. In vitro: In vivo protein-protein interactions represent true interactions within biological systems, while in vitro protein-protein interactions are recreated under controlled experimental conditions.
- Bimolecular Fluorescence Complementation (BiFC): Detects direct protein-protein interactions through fluorescence signals.
- Fluorescence Resonance Energy Transfer (FRET): Measures spatial proximity between proteins through energy transfer.
- Yeast Two-Hybrid (Y2H): Identifies directly interacting protein partners.
- Chromatin Immunoprecipitation Mass Spectrometry (ChIP-MS): Investigates protein-DNA interaction networks.
- Proximity Labeling Mass Spectrometry (PL-MS): Detects neighboring proteins in complex environments using a proximity labeling technique
- Co-immunoprecipitation (Co-IP): Captures protein complexes using specific antibodies, ideal for classic interaction studies.
- Affinity Purification Mass Spectrometry (AP-MS): Purifies interacting complexes using affinity tags for high specificity.
- Crosslinking Mass Spectrometry (XL-MS): Uses chemical crosslinkers to stabilize transient interactions and analyze their spatial relationships.
- Surface Plasmon Resonance (SPR): Monitors real-time interactions to assess binding kinetics and affinity.
- Isothermal Titration Calorimetry (ITC): Measures the thermodynamic properties and binding affinity between proteins.
- Disease Mechanism Analysis: Uncover pathogenic protein interaction networks and explore complex disease mechanisms.
- Drug Target Discovery: Identify interacting proteins associated with drug targets to guide drug development.
- Signal Pathway Research: Build signaling networks and analyze protein functions in biological processes.
- Biomarker Discovery: Identify potential biomarkers for diagnosis or prognosis, supporting precision medicine initiatives.
- Protein Function Research: Investigate protein interaction partners and their biological roles to advance basic research.
Protein-Protein interaction is essential for cellular functions and plays a central role in life sciences research by dynamically regulating biological processes such as signal transduction, gene expression, and metabolism. Understanding protein-protein interactions not only helps uncover protein functions but also provides crucial insights into drug development and disease mechanisms. MtoZ Biolabs offers professional and comprehensive protein-protein interaction service, leveraging state-of-the-art mass spectrometry platforms and a range of advanced techniques to support scientific discovery and accelerate research progress.
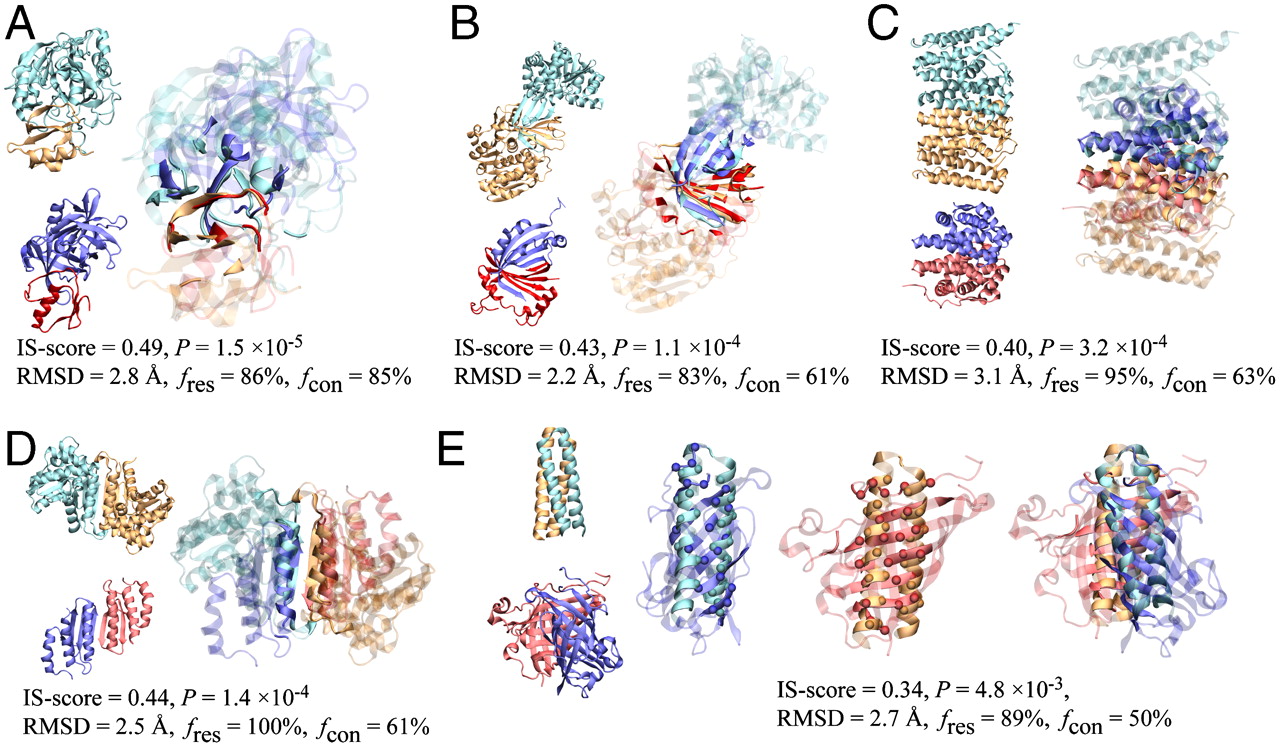
Gao, M. et al. PNAS. 2010.
Figure 1. Schematic Representation of Different Protein-Protein Interactions
Protein-Protein interactions can be classified based on several characteristics:
Services at MtoZ Biolabs
MtoZ Biolabs provides comprehensive protein-protein interaction analysis by integrating cutting-edge mass spectrometry technologies (e.g., LC-MS/MS) with a range of innovative methods. The protein-protein interaction service allows for the precise capture of target proteins and their interacting complexes, enabling us to analyze interaction networks and interpret their biological significance.
1. In Vivo Analytical Methods
In vivo protein-protein interaction service focuses on detecting interactions in their natural biological context, particularly those that are dynamic and functionally relevant. MtoZ Biolabs employs a range of techniques for precise analysis:
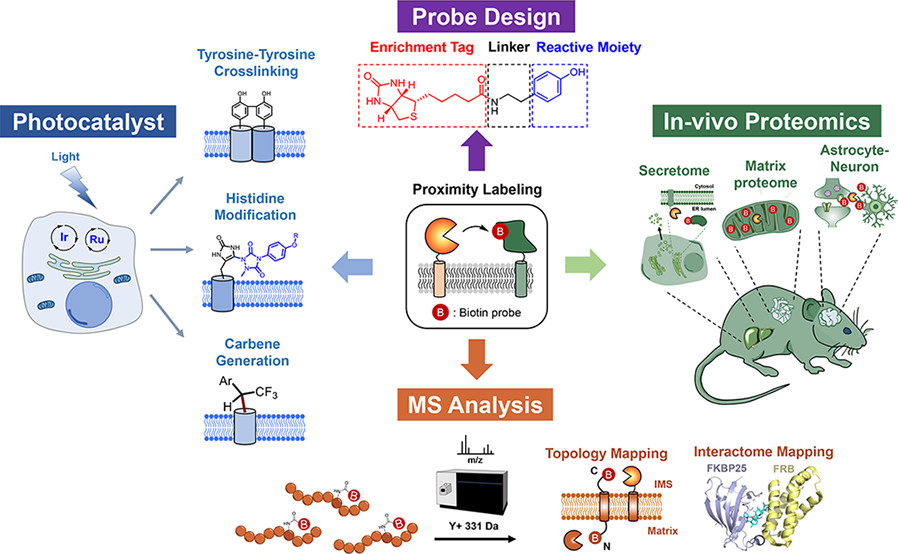
Kang, M. et al. Acc. Chem. Res. 2022.
Figure 2. The Workflow for PL-MS
These methods are ideal for exploring dynamic signaling pathways, inter-organelle interactions, and regulatory mechanisms of disease-related proteins.
2. In Vitro Analytical Methods
In vitro protein-protein interaction service involves optimizing experimental conditions to ensure high sensitivity and specificity. MtoZ Biolabs uses the following in vitro methods:
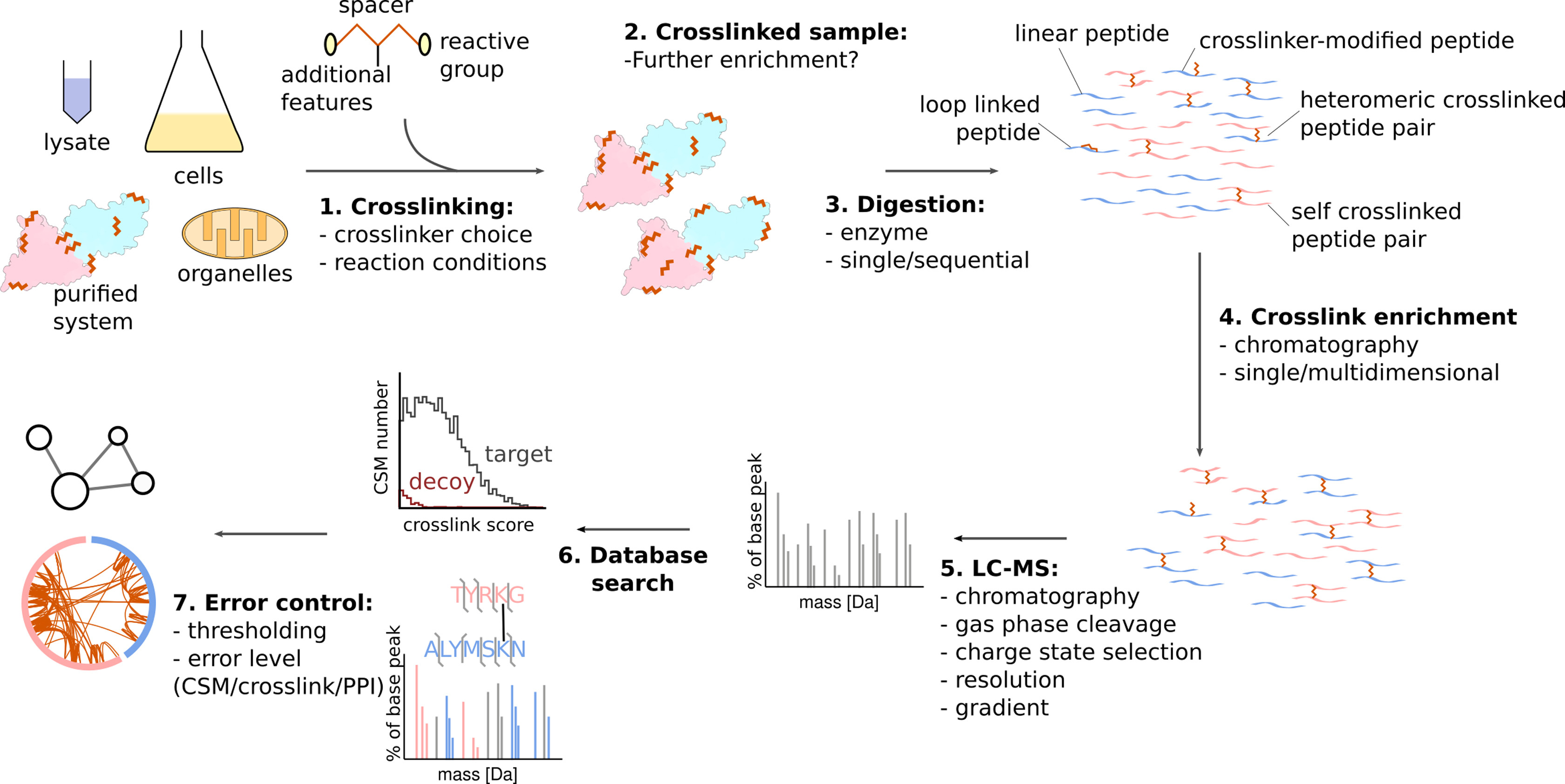
Graziadei, A. et al. Structure. 2022.
Figure 3. The Workflow for XL-MS
These techniques are highly suitable for validating direct or indirect interactions, and are widely applied in structural biology, complex research, and functional verification.
Service Advantages
1. Comprehensive Technical Support: We offer in vivo and in vitro analysis methods, providing full data support throughout the research process.
2. State-of-the-Art Equipment: Equipped with advanced mass spectrometers like the Thermo Orbitrap Fusion Lumos to ensure the highest data accuracy.
3. Customizable Solutions: We offer tailored services from experimental design to result interpretation, addressing diverse research needs.
4. High Data Integrity: By combining advanced mass spectrometry techniques with strict quality control, we ensure the precision and reproducibility of results.
5. Expert Team: Our experienced researchers ensure efficient project execution and progress.
Sample Submission Suggestions
1. Sample Types
Serum, plasma, urine, tissue extracts, and other biological samples.
2. Sample Volume
At least 200 μL of liquid sample or 200 mg of solid sample.
3. Sample Preservation
Store samples at -20°C or lower to prevent degradation, avoiding repeated freeze-thaw cycles.
Note: Provide details on sample collection and handling.
Applications
FAQ
Q1: Can protein-protein interaction service be used for membrane protein research?
Yes. Membrane proteins are challenging to study due to their hydrophobic nature. MtoZ Biolabs overcomes this by using optimized sample preparation protocols and advanced techniques like chemical crosslinking and affinity purification mass spectrometry to capture membrane proteins and their interacting partners. We also offer customized solutions for membrane protein studies.
Q2: How can we confirm if an interaction is real in a complex biological environment?
True interactions typically require both in vivo and in vitro verification. We recommend initial observations using in vivo techniques such as BiFC or FRET, followed by confirmation with in vitro methods like Co-IP or XL-MS. Cross-validation with independent experiments and multiple methods enhances result reliability.
Q3: Can protein-protein interaction analysis be combined with functional validation?
Yes. Our protein-protein interaction service is fully compatible with subsequent functional experiments. After identifying interacting proteins, you can use techniques like RNAi knockdown, overexpression, or mutation analysis to validate their roles in biological processes. We also offer functional proteomics services to comprehensively investigate protein functions.
What Could be Included in the Report?
1. Experimental Methodology and Workflow Overview
2. Sample Information and Processing Details
3. Mass Spectrometry Data and Quality Control Assessment
4. Data Preprocessing and Analysis Results
5. Bioinformatics Analysis and Interaction Network Mapping
6. Raw Data Files and Final Report
How to order?







