Proximity Proteomics Service
Proximity proteomics is a technique designed to investigate spatial protein-protein interactions within cells. Unlike traditional methods for detecting protein interactions, proximity proteomics uses a proximity labeling approach, allowing researchers to identify proteins that spatially interact with a target protein without requiring direct binding. This technology enables the discovery of novel protein interaction networks and provides deeper insights into complex biological processes such as cell signaling, metabolic regulation, and cytoskeletal dynamics.
MtoZ Biolabs offers advanced Proximity Proteomics Service to help researchers efficiently address critical questions about protein interactions and their biological functions. Our services are designed to precisely capture low-abundance, transient, or difficult-to-detect protein interactions, providing essential support for drug development, disease research, and biological mechanism studies.
Technical Principles
The core principle of proximity proteomics service is based on proximity labeling, in which enzymes like Biotin Ligase or TurboID trigger the tagging of nearby proteins, covalently attaching molecules (such as biotin) to proteins in close proximity to the target. This is achieved by genetically engineering the labeling enzyme to fuse with the target protein, ensuring that the enzyme is localized precisely in the region of interest. Once activated, the enzyme triggers a labeling reaction in a defined radius around the target protein, tagging both directly interacting proteins and those in the vicinity but not bound. The labeled proteins are then enriched using affinity purification techniques and analyzed through high-resolution mass spectrometry, enabling the construction of a detailed proximity interaction network for the target protein.
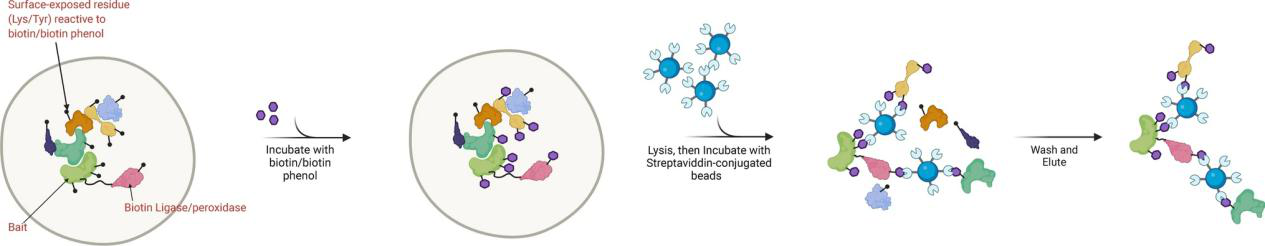
Low, T. Y. et al. Cell. Mol. Life Sci. 2021.
Figure 1. Proximity Proteomics Principle Diagram
Analysis Workflow
MtoZ Biolabs provides comprehensive Proximity Proteomics Service covering the entire workflow from gene tagging to data interpretation. Our advanced labeling techniques and high-resolution mass spectrometry platforms ensure efficient and accurate experimental outcomes.
1. Gene Insertion and Tagging
We fuse the target protein with a labeling enzyme, expressed in cells or tissues, to activate the labeling reaction and tag neighboring proteins.
2. Sample Processing and Protein Extraction
Cells or tissue samples are collected, lysed, and proteins, including the target and neighboring proteins, are extracted.
3. Enrichment of Labeled Proteins
Streptavidin beads are used to specifically capture the labeled protein complexes.
4. Mass Spectrometry Analysis
The labeled proteins are identified and quantified using high-resolution mass spectrometry techniques.
5. Data Analysis and Network Construction
Using bioinformatics tools, we analyze the mass spectrometry data to construct a detailed interaction network of the target protein.
Zhang, Y. L. et al. JOVE-J VIS EXP, 2020.
Service Advantages
Proximity Proteomics Service seamlessly integrates advanced detection capabilities with diverse research needs, providing a precise and versatile platform to explore complex protein interaction networks.
1. High Sensitivity: Accurately detects low-abundance, transient protein interactions that are challenging to capture with conventional techniques.
2. Unbiased Detection: No need for specific antibodies, offering comprehensive coverage of proteins interacting with the target.
3. Precise Enrichment: Efficiently captures labeled proteins, reducing background interference.
4. High-Resolution Analysis: Mass spectrometry allows for detailed identification and characterization of protein identities and modifications.
5. Wide Applicability: Applicable to a broad range of biological samples, including cells, tissues, plants, and clinical samples.
6. Customizable Solutions: Tailored experimental protocols to meet specific research needs.
Case Study
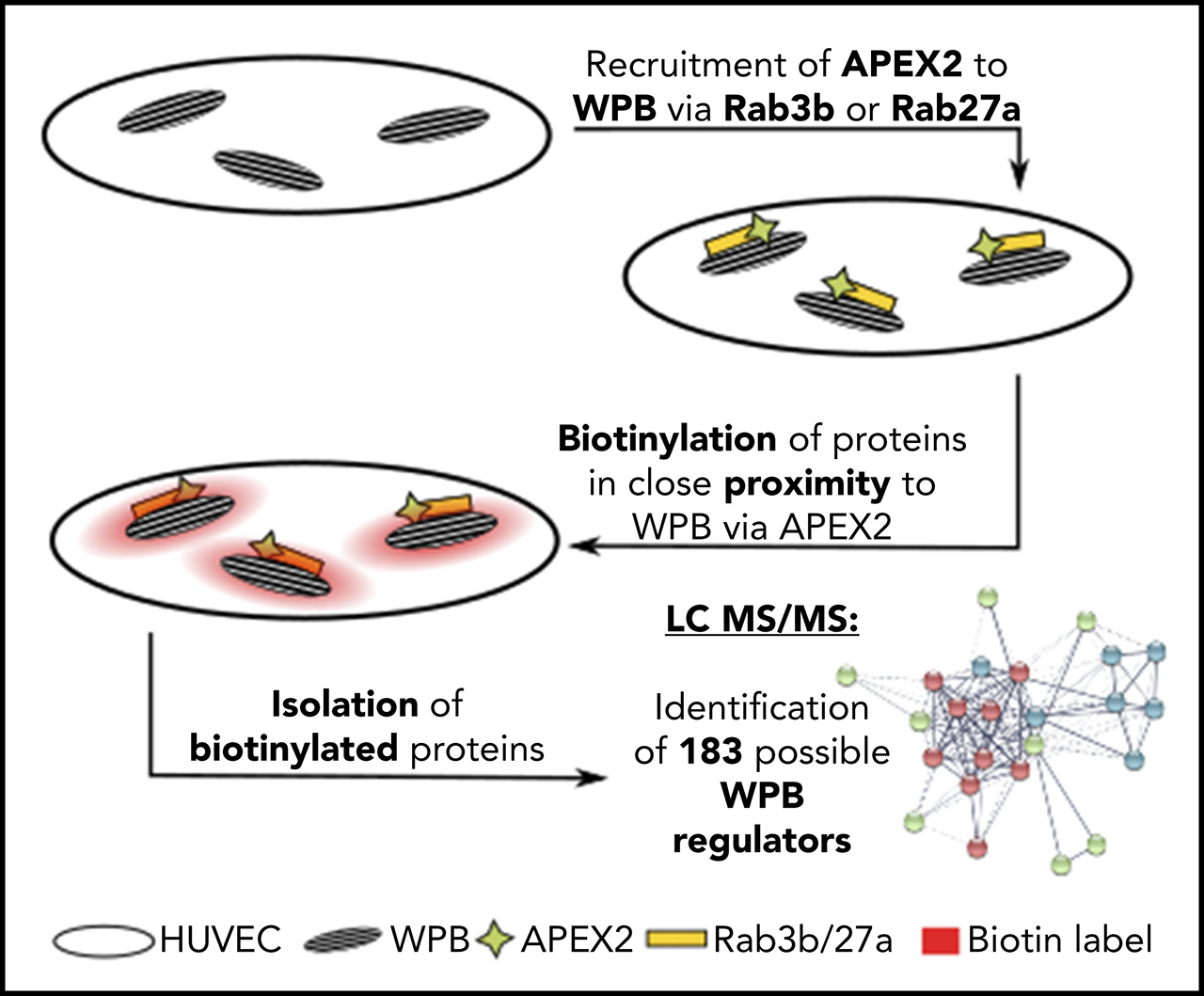
Case 1: Proximity Proteomics of the Weibel-Palade Body Protein Network
In this study, proximity proteomics was used to identify 183 proteins associated with Weibel-Palade bodies (WPBs), including several previously unknown proteins. Notably, the study found that the SNARE-interacting protein Munc13-2 specifically localizes to WPBs and regulates histamine-induced exocytosis and von Willebrand factor (VWF) secretion. This highlights the significant utility of proximity proteomics in elucidating the functions of cellular organelles and their protein networks.
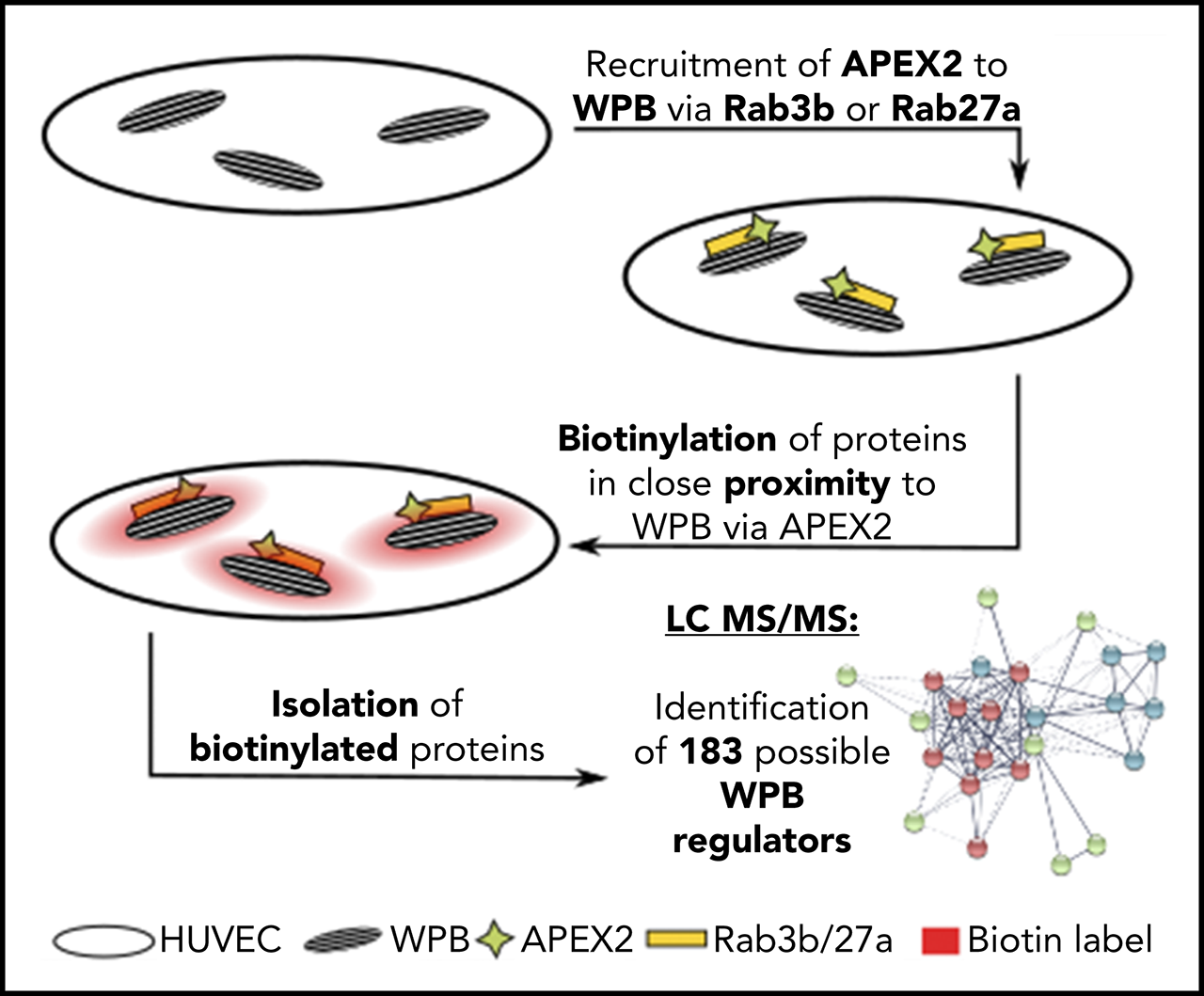
Holthenrich, A. et al. Blood. 2019.
Case 2: Proximity Proteomics Reveals the Role of PAK4 in Afadin–Nectin Junctions
In this study, proximity proteomics was applied to identify key neighboring proteins of PAK4 within the Afadin–Nectin cell junction complex. A total of 27 neighboring proteins and 17 phosphorylation sites were identified, providing new insights into the selective localization properties of PAK4 within these junctions. This research illustrates the power of proximity labeling and mass spectrometry in studying complex protein interaction networks and offers valuable insights into signaling mechanisms and potential disease targets.
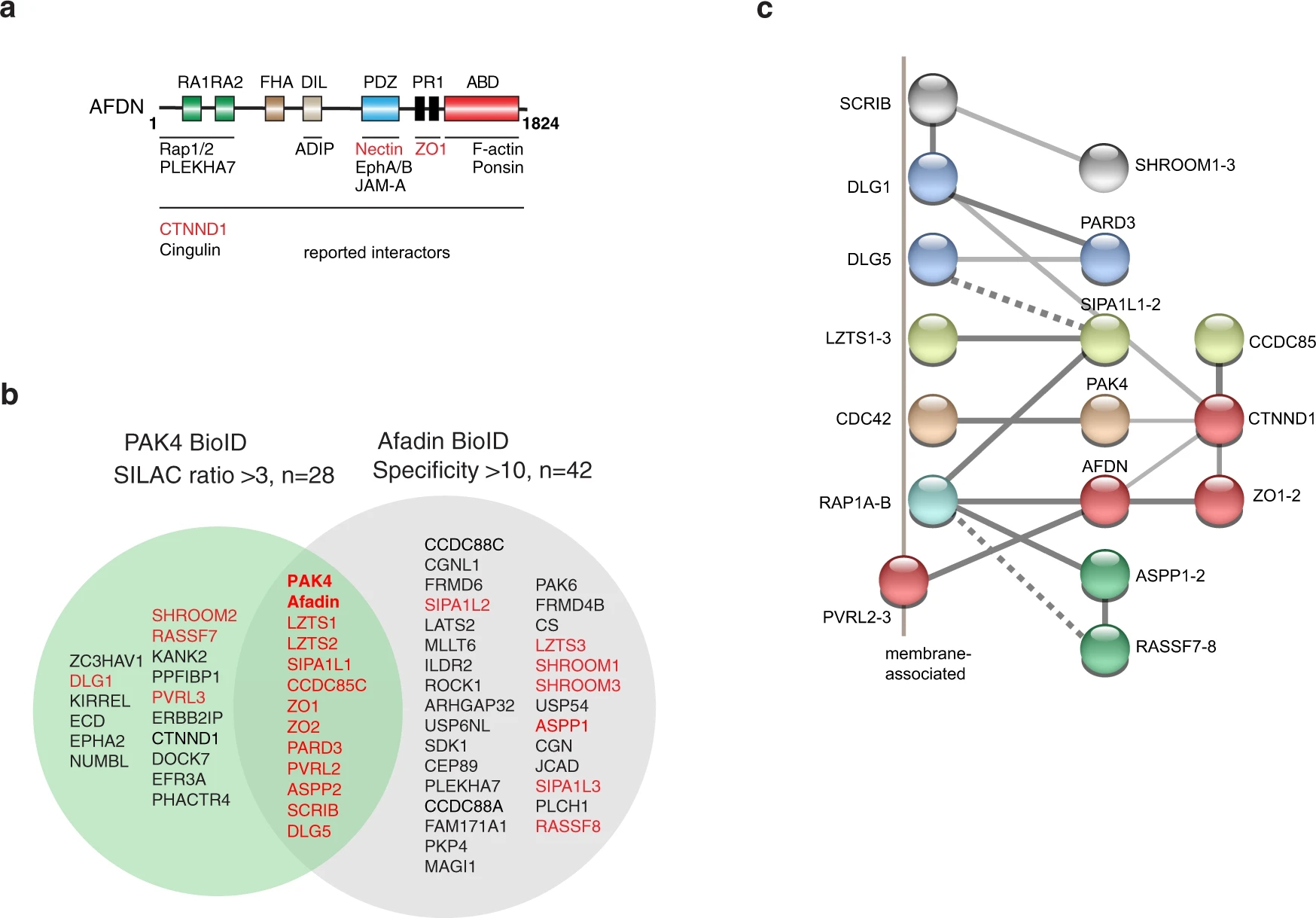
Baskaran, Y. et al. Nat. Commun. 2021.
Proximity proteomics can be broadly applied across various fields, including disease mechanism research, drug target discovery and validation, cell signaling pathway analysis, and protein complex dynamics. This technique is suitable for a wide range of biological sample types, including cells, tissues, plants, and clinical samples, and provides robust support for both basic scientific research and applied development.
At MtoZ Biolabs, we offer precise and efficient Proximity Proteomics Service designed to help you unravel complex protein interaction networks. With our cutting-edge technology platform and expert team, we are your trusted partner for advancing your research. Contact us today to begin a high-quality collaborative journey!
FAQ
Q1: What are the commonly used Proximity Proteomics tagging enzymes?
Commonly used tagging enzymes include BioID, APEX, and TurboID/miniTurboID. BioID is a Escherichia coli biotin ligase mutant that labels nearby lysines, requiring 18-24 hours with an optimal temperature of 37°C. APEX can rapidly label neighboring proteins within 1 minute but requires H2O2, which may induce cytotoxicity. TurboID and miniTurboID are highly efficient at labeling at room temperature, with a labeling time of about 10 minutes, and are especially suitable for plant systems. Based on your specific research objectives, our technical team can recommend the most appropriate enzyme for your study.
How to order?







