Comprehensive DNA Extraction and Purification Service
The sample preparation process encompasses: (1) releasing DNA from samples and cells through lysis, (2) isolating DNA, (3) purifying DNA by removing inhibitory substances, and (4) optionally, concentrating DNA, which is critical for detecting low-concentration analytes.
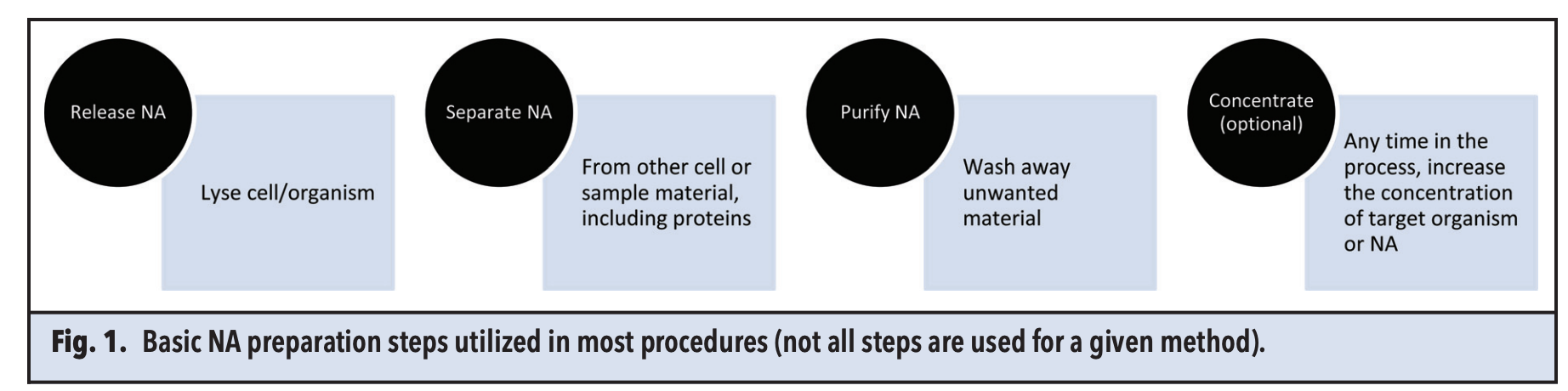
Figure 1. DNA Extraction Process [1]
1. Cell Lysis
DNA extraction commences with cell lysis to liberate DNA contents, employing physical and chemical lysis methods tailored to the sample type, such as cells, tissues, and plants.
(1) Physical Lysis
Physical Lysis methods include mechanical disruption, liquid homogenization, ultrasonic treatment, heating, freeze-thaw cycling processes, and manual grinding. Lysis poses a greater challenge when dealing with complex sample types or a wider variety of pathogens such as bacterial spores or ovary. These organisms require stronger chemical, enzymatic, or physical methods. Gram-positive bacteria have a thicker peptidoglycan layer that is difficult to digest and necessitates more extensive lysis. Bacterial spores, fungi, yeasts, and ovary have complex coats or walls containing cross-linked proteins and other complex molecules, enabling them to withstand many environmental factors, including chemical substances or biological defense systems. The disadvantages of physical methods of lysis include high costs, limited reproducibility, and localized heating within the sample.
(2) Chemical Lysis
Chemical lysis, including detergent-based lysis and enzymatic digestion using enzymes such as lysozyme and cellulase, serves as an effective alternative. Detergents such as SDS and Triton X-100 facilitate the separation of DNA from proteins. SDS is an early detergent used in DNA preparation, helping to separate DNA from nucleoproteins and all other protein sources, including membrane proteins. Another commonly used detergent is Triton X-100. Proteases are natural enzymes found in plants, animals, and microbes, and have a variety of uses. Some proteases are used in DNA preparations to reduce the protein background and assist in lysis by digesting membrane or coat proteins. Chemicals and detergents can only denature proteins, whereas proteases actually break them down into smaller molecules by cutting peptide bonds. The main proteases used in DNA preparations are serine proteases like Proteinase K. Although lysing samples under mild chemical conditions is advantageous, the lysis efficiency for certain cell types, such as plant cells, is low. The mixture obtained from ruptured membranes or walls contains a variety of soluble materials (such as proteins) and insoluble materials (such as cell fragments, like cell walls).
2. Nucleic Acid Extraction
DNA extraction techniques are classified into liquid-phase extraction (traditional) and solid-phase extraction based on the medium used.
(1) Liquid-Phase Extraction
Liquid-phase extraction involves purifying DNA after physically or chemically disrupting the sample, based on the different chemical properties of DNA compared to other components in the sample, through concentration, dissolution, and precipitation with the appropriate solvent. Main techniques include cesium chloride gradient centrifugation, alkaline lysis, and phenol-chloroform extraction.
DNA can be separated from other molecules by exploiting its differing solubility in immiscible liquids. The primary solvent used is phenol, typically mixed with chloroform and isoamyl alcohol. Phenol denatures proteins, which remain in the organic phase, while DNA is in the aqueous phase. The addition of chloroform and isoamyl alcohol helps separate the phases and prevent foaming. DNA in the aqueous phase is precipitated with ethanol to remove any residual phenol, obtaining a clean, concentrated product. Although this technique has wide molecular biology applications, it is cumbersome and time-consuming. Moreover, phenol and chloroform are susceptible to environmental contamination and may cause harm to researchers handling them. While not suited for high-throughput needs, microfluidic researchers still consider using it due to the ease of liquid manipulation.
DNA precipitation is a liquid-phase method used to obtain a very clean product and concentrate DNA. DNA is precipitated in the presence of alcohols (like ethanol or isopropanol) and high salt concentrations (like ammonium acetate, sodium acetate, or sodium chloride). Other liquids that can precipitate DNA include acetone and lithium chloride. Centrifugation concentrates the DNA precipitate from the remaining liquid, requiring manual operations to dry and resuspend the precipitate. This method produces very clean DNA but is tedious due to the manual handling involved.
(2) Solid-Phase Extraction
In contrast, solid-phase extraction utilizes the interactions between solid adsorbents and DNA (electrostatic, affinity, iocmite, glass fiber, and anion-exchange carriers) to achieve separation. Solid-phase extraction is a standard method for nucleic acid extraction, primarily represented by silica-based spin columns and silica-based magnetic bead methods.
Silica gel-based spin column DNA extraction was first developed in 1979. DNA in the lysate specifically adsorbs to the surface of a silica-based membrane under high salt conditions, while remaining impurities in the lysate are washed away through multiple washing steps, followed by a final elution to obtain the extracted DNA. A major drawback of the spin column method is the low adsorption efficiency of certain columns for DNA. The silica gel membranes used for DNA separation have the disadvantage of low binding capacity for nucleic acids, resulting in relatively less extracted DNA.
Compared to traditional methods, the magnetic bead method is relatively feasible, simple, fast, and capable of high-throughput operations. The principle of the magnetic bead is similar to that of the spin column method; magnetic beads specifically adsorb DNA, which is then cleansed of other cell lysate components through a series of washing processes. In existing technology, magnetic nanoparticles typically have a coating. Silica (SiO₂) is widely used as a coating for magnetic nanoparticles due to its good hydrophilicity, non-toxicity, and protective properties. Thus, the target DNA is specifically adsorbed onto the SiO₂ layer on the magnetic beads, facilitating the separation and purification of DNA. Although SiO₂-coated magnetic nanoparticles can extract nucleic acid molecules, like the spin column method, these nanoparticles generally suffer from low adsorption efficiency, often failing to achieve efficient nucleic acid extraction. High costs, the non-recoverability of magnetic beads, and potential aggregability are drawbacks of the magnetic bead method.
The paper surface binding method, where DNA binds with cellulose, is a convenient and quick DNA preparation method. Chemically treated paper contains lysing and binding agents that combine with the sample. Lysis and binding occur in the paper, then washed and eluted from the paper, typically involving small volumes.
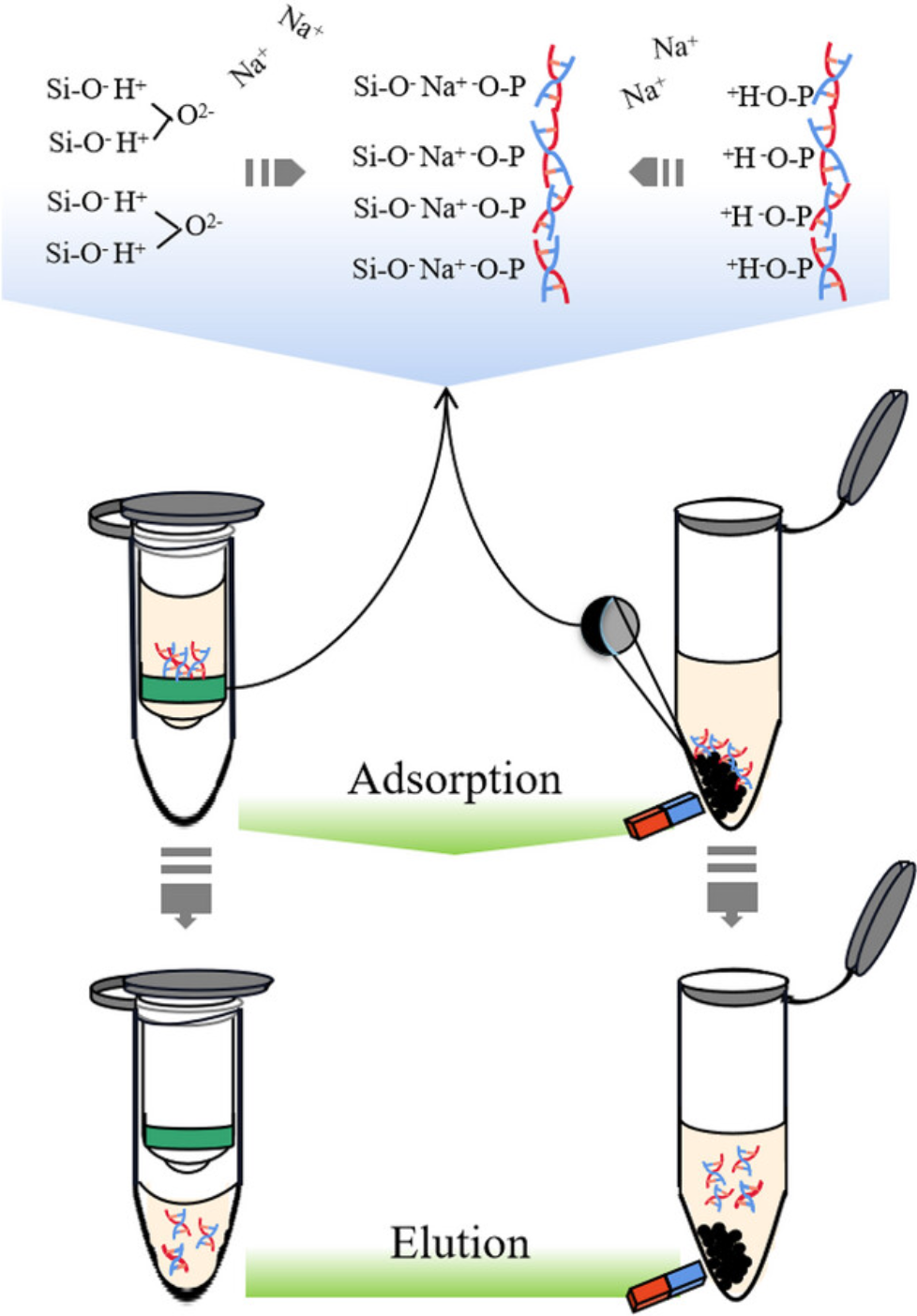
Figure 2. Principle of DNA Extraction by Spin Column and Magnetic Beads Method [2]
3. DNA Sample Purification, Decontamination, and Quality Control
During the nucleic acid extraction process, there are two potential sources of contamination (downstream inhibitors or impurities): the reagents used in the extraction process and the original samples. To address these issues, current nucleic acid extraction techniques still need further development. Most impurities introduced during the extraction process are chemical substances that interfere with downstream reactions, known as inhibitors. It is important to note that the concentration of these inhibitors is more critical than their characteristics in terms of interference with downstream detection. Based on the elution solutions of four major commercial kits, it has been demonstrated that inhibitors (such as PCR LAMP) have weak interference at high dilutions, while at low dilutions, downstream reactions are significantly inhibited. Since inhibitors and extracted nucleic acids coexist in the elution solution, diluting the inhibitors to minimize interference also dilutes the extracted nucleic acids, which is a drawback, especially when the target DNA is scarce and/or high sensitivity is required (e.g., for cell-free DNA [cfDNA], single-cell, and single-nucleotide polymorphism analysis). To obtain concentrated nucleic acid samples and eliminate the interference of inhibitors, a method called two-phase elution has been developed, which involves incorporating a hydrophobic buffer prior to elution, to facilitate testing of low-dilution samples.
The purity of extracted nucleic acids can be accurately assessed using OD260/OD280 and OD260/OD230 ratios. However, most related studies only provide the OD260/OD280 ratios for extracted DNA, while the OD260/OD230 ratio is generally overlooked. Relying solely on the OD260/OD280 ratio is insufficient to estimate the purity of extracted nucleic acids because nucleic acids absorb strongly at OD260, and only high concentrations of protein can be detected by the OD260/OD280 ratio. However, the OD260/OD230 ratio also provides critical purity data, especially for downstream analysis that requires high purity nucleic acids. Protein residues cause a significant increase in OD230, significantly affecting the OD260/OD230 ratio. Specifically, for DNA, an OD260/OD280 ratio greater than 2.0 indicates RNA contamination, while a ratio less than 1.7 indicates the presence of proteins and phenolic compounds; additionally, an OD260/OD230 ratio less than 2.0 indicates the presence of polysaccharides, salts, or other organic solvent residues. Therefore, a suitable extracted DNA's OD260/OD280 ratio should be between 1.7 and 2.0, and the OD260/OD230 ratio should be higher than 2.0. Therefore, it is not recommended to overlook purity assessments when evaluating extraction efficiency.
Analysis Workflow
1. Experimental Procedure Determination Based on Experimental Needs
2. Cell Lysis
3. DNA Extraction
4. DNA Quality Control
Service Advantages
1. High Credibility Identification and Characterization of Host Cell Residual DNA
2. Automated High-Throughput DNA Extraction Process
3. Effective DNA Detection System and Extensive Testing Experience
Sample Results
1. DNA Extraction Using TRIzol Reagent with Silica Columns
TRIzol is a single-phase solution of phenol and guanidinium isothiocyanate, used for extracting RNA, DNA, and proteins from tissues or cells. However, due to its time-consuming process, few studies have described its application in DNA extraction. Research has proposed a TRIzol-modified method using TRIzol reagent and silica columns for DNA extraction from tissues, which requires only one-third the time of the classical extraction procedure. Spectrophotometric analysis shows that the 260/280 and 260/230 nm absorbance ratios of DNA extracted using the TRIzol-modified method are ideal, comparable to those obtained with the classical method and commercial DNAiso methods. The performance of DNA extracted using the TRIzol-modified method in restriction enzyme digestion and quantitative PCR is the same as that of DNA extracted using the classical method. The TRIzol-modified method saves time, simplifies the DNA extraction procedure, and facilitates various molecular biological analyses.
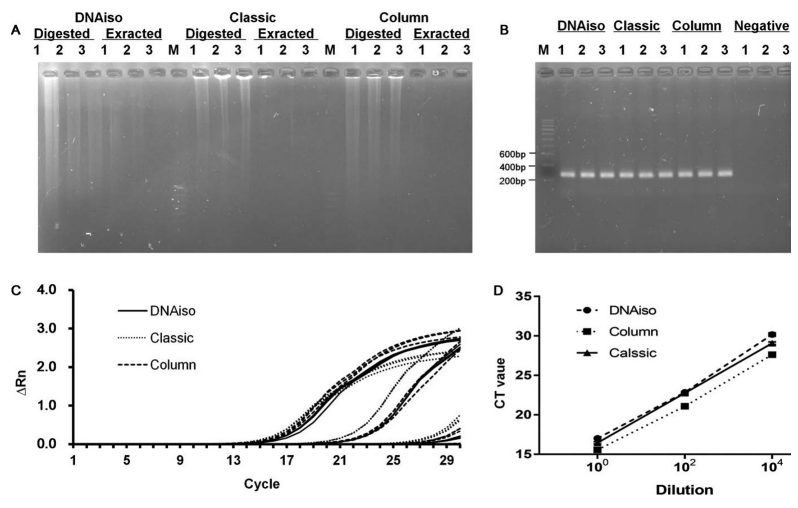
Figure 3. DNA Extraction Effect of Different Methods [3]
2. Magnetic Ionic Liquid DNA Extraction: Adjustable Solvents for Rapid and Selective DNA Analysis
DNA extraction is a critical bottleneck in nucleic acid analysis. Research has synthesized hydrophobic magnetic ionic liquids (MILs) and used them as solvents for the rapid and efficient extraction of DNA from aqueous solutions. DNA-rich droplets can be manipulated by applying a magnetic field. The three MILs studied demonstrated unique DNA extraction capabilities when applied to various DNA samples and matrices. Using benzyltrioctylammonium bromotrichloroferrate(III) ([(C 8)3 BnN +] [FeCl 3Br -]) was highly efficient for extracting smaller single-stranded and double-stranded DNA. On the other hand, the dicationic 1,12-di(3-hexadecyl benzimidazolium) dodecane bis[(trifluoromethyl) sulfonyl]imide bromotrichloroferrate(III) ([(C16BnIM)2C122+][NTf2-, FeCl3Br-]) was more efficient for extracting larger DNA molecules. The MIL-based method was also used to extract DNA from complex matrices containing albumin, indicating that trihexyl(tetradecyl)phosphonium tetrachloroferrate(III) ([P6,6,6,14+][FeCl4-]) exhibits competitive extraction behavior. Compared with [(C8)3BnN+][FeCl3Br-4] MIL, this MIL significantly reduced the co-extraction of albumin. The MIL-DNA method was employed for extracting plasmid DNA from bacterial cell lysates. The quality and quantity of DNA recovered from the MIL extraction phase were sufficient for polymerase chain reaction (PCR) amplification, demonstrating the feasibility of MIL-based DNA sample preparation before downstream analysis.
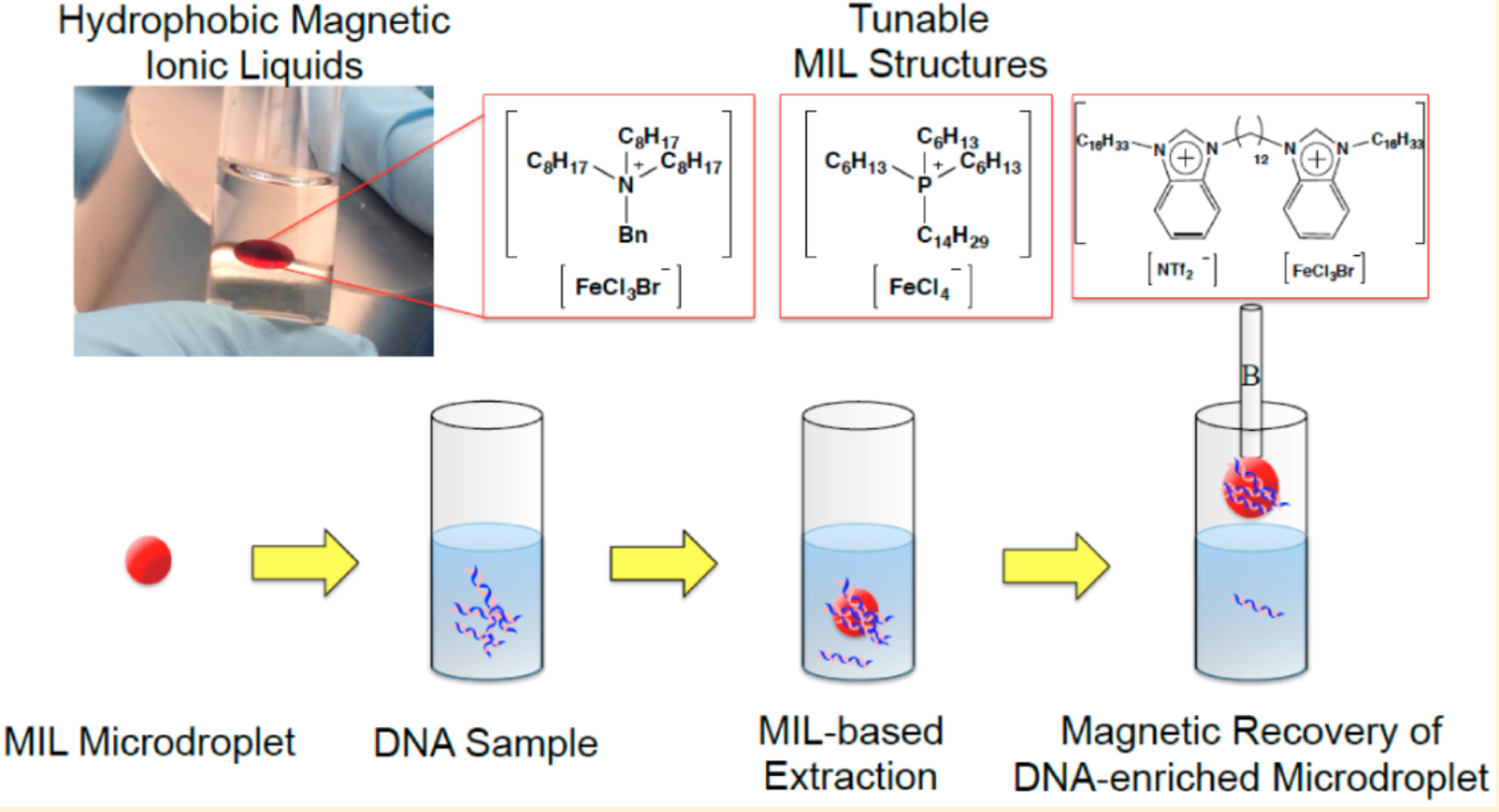
Figure 4. Method for DNA Extraction Based on MIL [4]
3. Integrated PCR Device Based on Cellulose Membrane
Research has reported the design, fabrication, and testing of a microfluidic device for molecular diagnostics that can complete lysis, solid-phase nucleic acid extraction, polymerase chain reaction (PCR) amplification, and real-time fluorescence detection all within a single chamber. This streamlined design greatly simplifies the manufacturing and operation of chip-based nucleic acid detection for pathogens and other disease markers. The single 25 µL PCR chamber in the plastic chip is equipped with a cellulose-based filter membrane (Whatman FTA®), used for nucleic acid extraction from samples. The nucleic acids captured on the membrane serve as templates for in-situ PCR amplification. The disposable microfluidic chip is used with a portable instrument that heats the PCR chamber bilaterally for rapid thermal cycling and employs a low-cost, miniaturized (ESE GmbH) fluorescence reader for real-time fluorescence detection. The study optimized multiple sample loading on the FTA® membrane, vacuum drying of the membrane, and membrane washing steps to enhance extraction efficiency in a microfluidic format. The chip was tested with Bacillus cereus bacterial culture samples, demonstrating a detection limit of ~103 target cells. The paper discusses the adaptability of the chip in practical microfluidic applications, including integrated bags for storage and fluid driving, pre-loaded dry-stored PCR reagents, and potential near-term applications.
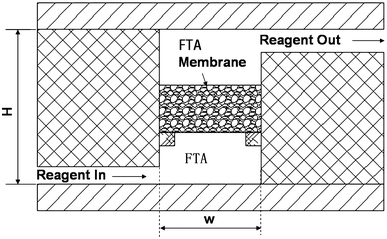
Figure 5. PCR Reactor Equipped with Circulating FTA Membrane [5]
4. Portable Integrated Microfluidic Technology for Rapid and Sensitive Diagnosis of Streptococcus Agalactiae in Resource-Limited Settings
Streptococcus agalactiae (Group B Streptococci, GBS) remains a significant neonatal infection source, making rapid and accurate GBS diagnosis in pregnant women crucial to preventing infections. Traditional nucleic acid amplification-based detection methods have been applied in central laboratories for GBS diagnosis, demonstrating high sensitivity. However, these methods heavily rely on equipment and trained technicians, posing significant barriers to widespread GBS testing, including self-testing and bedside/community screening. Moreover, the structure of GBS presents additional challenges for nucleic acid extraction and purification. A novel GBS diagnostic platform that integrates sample processing, amplification, and readout is highly anticipated in clinical settings. A study reported a portable integrated microfluidic technology that enables rapid DNA extraction (<10 minutes) from sampling swabs, power-free DNA amplification (<30 minutes), and simple readout for GBS detection. This platform operates without external pumps, achieving rapid and efficient DNA extraction from clinical samples, significantly reducing the time from 6 hours to under 50 minutes. Systematic clinical testing based on 47 patient samples confirmed the high performance of the platform, particularly its low detection limit (LOD, 103 copies/mL), high sensitivity (100%), and high specificity (100%). A head-to-head comparison showed that this device's LOD improved by an order of magnitude compared to traditional PCR methods, providing a simple yet powerful POCT platform for home/community testing of GBS (and other pathogens) in remote areas.
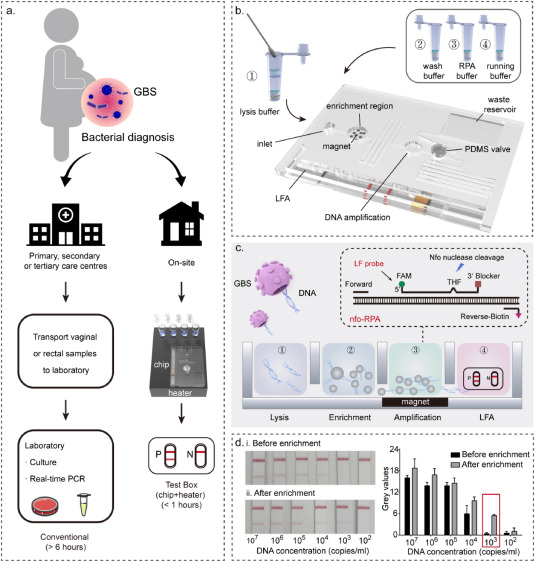
Figure 6. Design Concept and Operation Flow of PIM for GBS Rapid Diagnosis [6]
Sample Submission Requirements
1.Minimize Impurity Contamination
Services at MtoZ Biolabs
1. Complete Experimental Procedure
2. Relevant Instrument Parameters
3. Original Experimental Data
4. Data Analysis Submission
Applications
1. Forensic DNA Extraction From Human Tissues
DNA identification of human remains plays a crucial role in forensic science and even more broadly. Despite DNA being vital for identifying unknown human remains, post-mortem environmental factors can lead to poor molecular preservation. In this regard, the focus is on DNA extraction methods from hard tissue samples, as these samples have the longest survival times. Despite decades of research into DNA extraction methods from bones and teeth, there is almost no consensus on optimal performance. Results from meta-analyses have shown that solid-phase magnetic bead/resin extraction techniques are associated with higher DNA profiling success rates, and adding a demineralization step before extraction can improve the success rate of Short Tandem Repeat (STR) profiling.
2. Environmental DNA for Wildlife Biology and Biodiversity Monitoring
Extracting and identifying DNA from environmental samples has recently achieved significant success in detecting and monitoring common, endangered, invasive, or elusive species. The special attributes of environmental DNA (eDNA) analysis make it a powerful tool for elucidating the mechanisms of ecological and evolutionary processes. Most importantly, it enhances the ability to explore ecosystem-level processes, providing quantitative measures for species, community diversity, and dynamics analysis, and offering new opportunities for detecting rare or difficult-to-sample taxa through the use of time series samples and unprecedented sensitivity.
References
[1] Thatcher SA. DNA/RNA preparation for molecular detection. Clin Chem. 2015 Jan;61(1):89-99. doi: 10.1373/clinchem.2014.221374. Epub 2014 Dec 1. PMID: 25451869.
[2] Ye X, Lei B. The current status and trends of DNA extraction. Bioessays. 2023 Aug;45(8):e2200242. doi: 10.1002/bies.202200242. Epub 2023 Jun 20. PMID: 37338306.
[3] Yang BH, Liu BS, Chen ZL. DNA Extraction with TRIzol Reagent Using a Silica Column. Anal Sci. 2021 Jul 10;37(7):1033-1037. doi: 10.2116/analsci.20P361. Epub 2020 Nov 27. PMID: 33250452.
[4] Clark KD, Nacham O, Yu H, Li T, Yamsek MM, Ronning DR, Anderson JL. Extraction of DNA by magnetic ionic liquids: tunable solvents for rapid and selective DNA analysis. Anal Chem. 2015 Feb 3;87(3):1552-9. doi: 10.1021/ac504260t. Epub 2015 Jan 22. PMID: 25582771.
[5] Qiu, X., Mauk, M.G. An integrated, cellulose membrane-based PCR chamber. Microsyst Technol 21, 841–850 (2015). https://doi.org/10.1007/s00542-014-2123-x.
[6] Wang Z, Yan B, Ni Y, Cao Y, Qiu J, He R, Dong Y, Hao M, Wang W, Wang C, Su H, Yi B, Chang L. A portable, integrated microfluidics for rapid and sensitive diagnosis of Streptococcus agalactiae in resource-limited environments. Biosens Bioelectron. 2023 Dec 9;247:115917. doi: 10.1016/j.bios.2023.115917. Epub ahead of print. PMID: 38101186.
How to order?







