Tumor-Associated Antigens 1 Screening Service
T cells play a vital role in destroying pathogens and monitoring pathological cells. They are able to distinguish between self-antigens and non-self-antigens by surface T cell receptors (TCRs). Most human T cells express the classical TCRs, which consists of TCRα and TCRβ chains co-expressed with CD3 chains (γ, δ, ε, and ζ). The TCRα and TCRβ chains are composed of a variable (V) and a constant (C), respectively. The Vα chain is encoded by V and joining (J) gene segments (TRAV and TRAJ), while the Vβ chain is composed of V, diversity (D), and J gene fragments (TRBV, TRBD, and TRBJ). Three complementarity-determining regions (CDRs) located within the TCR V domain are the main parts of the TCR's interaction with the peptide-MHC (pMHC) complex, thus conferring TCR antigen specificity. Recent results from high-throughput sequencing (HTS) suggest that the actual diversity of the TCR gene pool may be between 1011-1012 unique TCRs. Upon recognition of the homologous pMHC complex by TCR, the immunoreceptor tyrosine-based activation motif (ITAM) in the CD3 chain is immediately phosphorylated, triggering a signaling cascade that leads to T cell activation and induces T cell function.
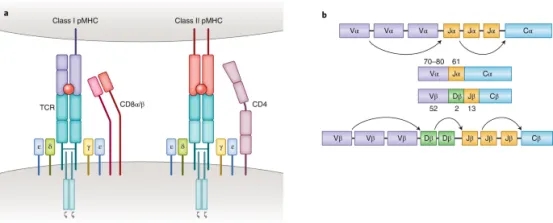
Figure 1. Molecular Basis of TCR-pMHC Recognition [2]
TCR-pMHC interaction is a key factor in adaptive immunity. Classical CD8+ and CD4+ αβΤ cells recognize short peptides presented by the MHC class I and II (MHC I and II) molecules, respectively. In contrast to these MHC-restricted αβ T cells, unconventional γδT cells composed of TCRγ and TCRδ chains are able to recognize non-peptide antigens (e.g., lipid antigens) in an MHC-unrestricted manner. Another characteristic of TCR-pMHC interactions is promiscuity: each TCR is capable of identifying millions of different peptides, and many different TCRs can recognize a given peptide.
The identification and selection of tumor-associated antigens (TAAs) or tumor neoantigens for clinical applications requires more thorough validation rather than simple confirmation as tumor-specific targets. The ideal therapeutic target should have the highest levels of tumor specificity, human leukocyte antigen (HLA) binding capacity, and immunogenicity. Antigens lacking any of these prerequisites, including those that induce T-cell incompetence or tolerance, are not only not therapeutically beneficial, but may even be harmful. Therefore, a series of conditional screening of TAAs and neoantigens is of great significance for the safe development of subsequent treatments.
1. Overview of Antigen-Directed Methods
To elucidate antigen-specific T cell responses, various antigen-directed methodologies have been established. A principal approach involves the assessment of TCR-pMHC interactions by evaluating T cells' functional response upon exposure to specific antigens. Key techniques encompass T cell proliferation measurement, antigen-specific cytolytic activity via chromium release assays, enzyme-linked immunospot (ELISpot) assays, and intracellular cytokine staining. A significant challenge across these methods is the requisite for a substantial number of TCR-expressing T cells.
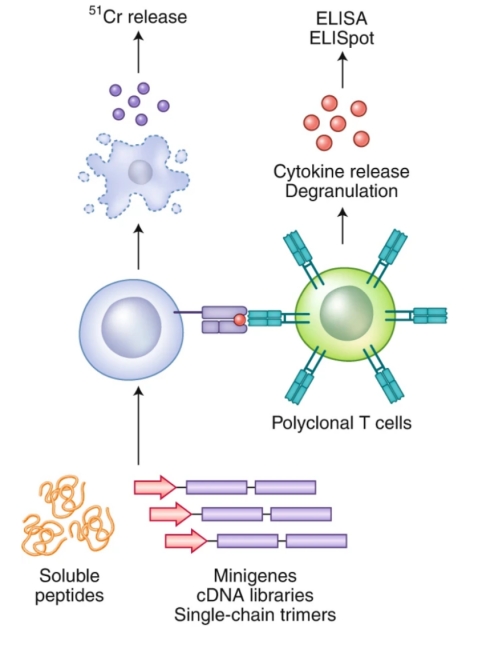
Figure 2. Illustration of TCR-pMHC Interaction Characterization through Cytokine Release and/or Cytotoxicity Analysis [2]
(1) ELISpot Assay
Recognized for its sensitivity within cellular immunology, the ELISpot assay facilitates detection of both antibody-secreting (B cells) and cytokine-secreting (T cells) at the single-cell level. Capable of identifying a single secreting cell amongst 200,000 to 300,000 cells, this assay triggers immune cell activation in the presence of stimulants. Secretions are captured by specific monoclonal antibodies (mAbs) affixed to the ELISpot plate's PVDF membrane. Following cell removal, captured cytokines bind to biotin-labeled mAbs, then to enzyme-linked avidin, and develop color upon substrate addition. Resultant spots signify cytokine production, quantified through an ELISpot Reader.
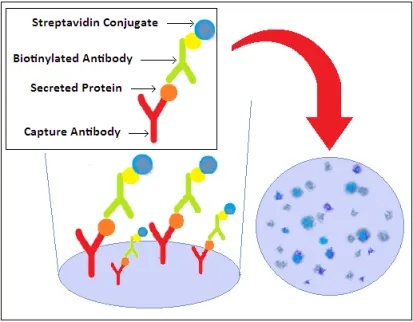
Figure 3. ELISpot Assay Principle
(2) Detection of T Cell Activation
The activation of naive T cells into CD4+ helper and CD8+ cytotoxic T cell clones is driven by antigens and co-stimulatory signals. Beyond proliferation, this activation initiates multiple signaling pathways, altering functional cell surface markers and cytokine release. Analysis typically involves measuring the expression of CD69, CD25, and HLA-DR activation markers. CD137 and other immune checkpoints on T cells further denote activation levels.
(3) Monitoring T Cell Proliferation
Flow cytometry serves as a standard for tracking cell proliferation, utilizing dyes like CFSE and PKH26 to mark living cells. These dyes dilute upon cell division, allowing subsequent generations to be distinguished by fluorescence intensity reduction. Peptide libraries tailored to experimental needs are synthesized, with donor PBMCs tagged using CFSE. Co-culturing with peptides elicits CD4+ T cell proliferation, observable through CFSE dilution and analyzed via flow cytometry to gauge antigen response.
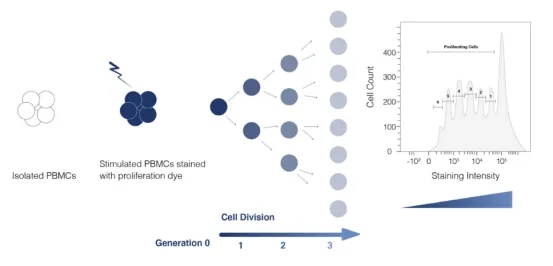
Figure 4. Principle of Flow Cytometric Detection of Cell Proliferation
(4) Assessment of T Cell-mediated Cytotoxicity
Evaluating T cells' cytotoxicity towards target cells employs methods like the LDH assay, where lactate dehydrogenase release indicates cell membrane compromise. Additionally, reporter gene transfection techniques gauge cytotoxic effects by integrating reporter enzymes (e.g., β-galactosidase, luciferase) into target cells. Analyzing these enzymes' activity in the culture medium reflects the extent of target cell death, thus quantifying the effectiveness of T cell-mediated killing.
2. Strategies for TCR-Directed Screening
The overarching strategy in TCR-directed approaches is to sift through extensive libraries of pMHC complexes for specific TCRs or T cell clones. Initial methods involved combinatorial peptide libraries that randomized all but one position, aiming to isolate clones of interest.
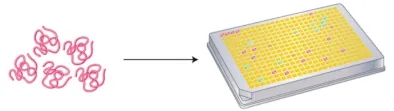
Figure 5. Utilization of a Combinatorial Peptide Library with Randomized Amino Acids for Functional Assays Targeting Epitope Screening[2]
Peptides can also be engineered genetically and presented on the surfaces of bacterial, yeast, or mammalian cells as part of pMHC complexes. A significant advantage of using yeast display is its ability to initiate with a vast, truly diverse library of billions of pMHC complexes, eliminating the need for prior epitope knowledge. Its primary challenge lies in the necessity to modify MHC alleles to improve folding, stability, and pMHC presentation adequately. Unlike yeast display, phage display does not require these modifications. Despite yeast display predominantly generating mimotopes, recent advances in algorithms have substantially enhanced the mapping of these mimotopes to their corresponding natural peptides. A common limitation of both yeast and phage display techniques is the requirement for generating soluble TCRs, a process known for its instability. These display techniques involve presenting a peptide library within Sf9 insect or yeast cells, staining with fluorescently labeled soluble TCRs, followed by FACS sorting of the labeled target cells for sequencing and epitope determination. The NNK codon scheme is employed, representing degenerate codons where "N" stands for any nucleotide, and "K" represents either guanine (G) or thymine (T).
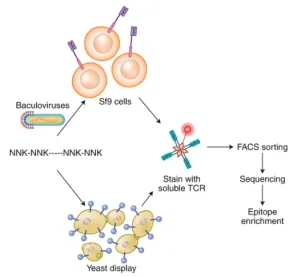
Figure 6. Overview of Yeast and Phage Display Techniques for Epitope Screening [2]
Contrasting with the passive screening methodologies used in yeast or phage displays, mammalian cell surface display techniques introduce an active screening mechanism. This method distinguishes homologous pMHC complexes from unrelated ones by producing a specific signal. This innovative approach utilizes Signal and signaling and antigen-presenting bifunctional receptors (SABRs), which are fusion molecules combining extracellular pMHC complexes with the intracellular signaling domains of CD28-CD3ζ. Upon TCR recognition, SABRs signal to antigen-presenting cells (APCs). Utilizing NFAT-GFP Jurkat cells, which express GFP in response to CD28-CD3ζ signaling, allows the identification of SABRs recognized by particular TCRs.
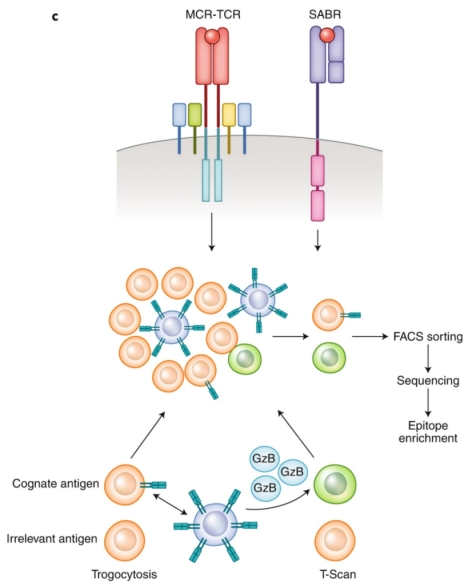
Figure 7. The SABRs Technique for Discriminative Screening of Target Epitopes [2]
3. Bioinformatics-Driven Screening
The evolution of high-throughput TCR sequencing and the accumulation of specificity data between TCR-pMHC have catalyzed the development of algorithms, paving the way for computational antigen discovery. These methodologies rest on the premise that TCRs with similar CDR3 motifs may share antigenic specificities. Groundbreaking studies in 2017 by Glanville et al. demonstrated the feasibility of discovering antigens computationally. They proposed that certain CDR3 motifs correlate with specific antigenic specificities. Analyzing 266,436 unique TCR sequences, they assessed the enrichment and probability of enrichment for various 2-, 3-, and 4-residue motifs, revealing clusters of TCR sequences with high similarity within antigen-specific libraries. The grouping of lymphocyte interactions by paratope hotspots (GLIPH) algorithm emerged from this work, clustering TCRs by potential common targets based on sequence similarity, structural insights, V gene usage, CDR3 length variability, and HLA typing. This body of research underscores that TCRs recognizing the same antigen often share sequence and structural characteristics, allowing for their classification based on these features. By reversing this classification process, it becomes possible to group TCRs into categories predictive of their antigenic specificities, incorporating considerations of force fields, structural data, or the biochemical properties of amino acids.
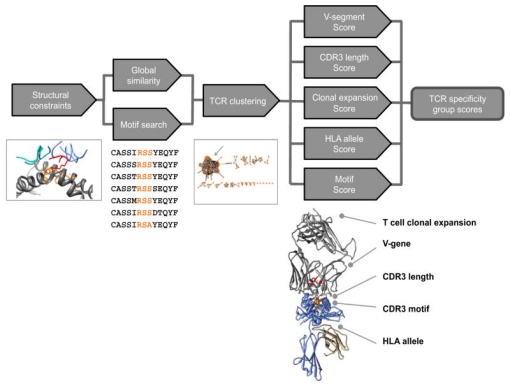
Figure 8. The GLIPH Algorithm's Three-step Process for Antigen Discovery [3]
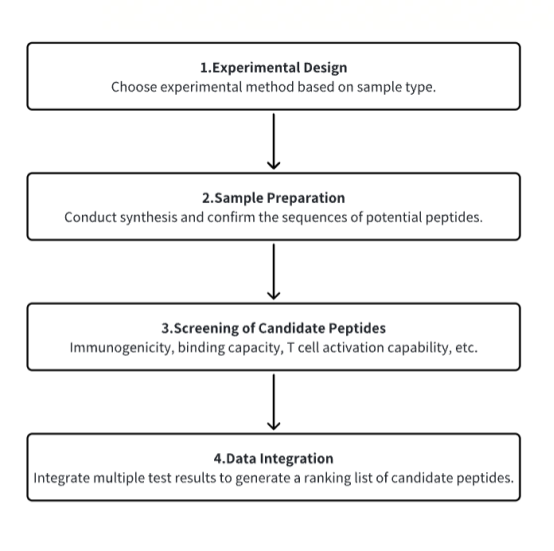
Analysis Workflow
1. Choose methods tailored to the specific needs of the experiment.
2. Conduct synthesis and confirm the sequences of potential peptides.
3. Evaluate peptides for immunogenicity, binding capabilities, and the ability to activate T cells, among other criteria.
4. Compile and analyze the collected data for insights.
Service Advantages
1. Tailor experiments to meet specific research requirements.
2. Utilize a high-throughput platform for efficient screening of antigenic peptides.
3. Employ detailed bioinformatics tools for in-depth analysis.
Sample Results
1. Immunogenicity and Targeting of Neoantigens from Mutated PIK3CA
Neoantigens, derived from recurrent mutations in oncogenes like PIK3CA, represent shared, cancer-specific epitopes. A novel high-throughput methodology integrating single-cell transcriptomics with TCR sequencing was employed to assess if PIK3CA mutations yield immunogenic neoantigens. This led to identifying TCRs that recognize a peptide incorporating a frequent PIK3CA mutation, presented by the common HLA-A*03:01 allele. This neoantigen's immunogenicity is attributed to enhanced peptide/HLA complex stability, due to HLA’s preference for certain anchor residues. Structural analyses revealed the bound peptide presents a largely featureless surface, emphasizing the peptide backbone. A leading TCR candidate demonstrated high specificity and affinity, attributed to an extended binding interface facilitated by an unusually long CDR3β loop. Observations across various tumors highlighted the neoantigen's immunogenicity, with specific TCR-engineered T cells inducing regression in PIK3CA-mutant tumors in vivo, underscoring the potential for therapeutic applications.
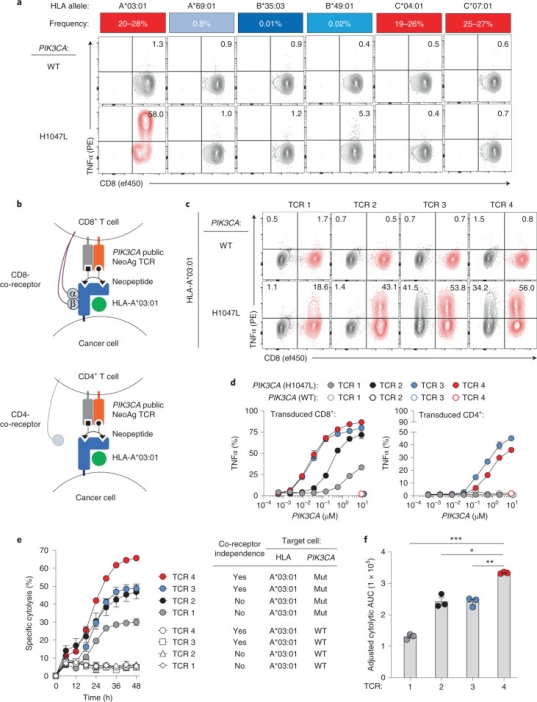
Figure 9. Functional Characterization and HLA Restriction of PIK3CA-specific TCRs [4]
2. High-Throughput Neoantigen and TCR Screening Using Droplet Microfluidics and Jurkat Reporter System
Engineered T cells with TCRs targeting a wide array of tumor antigens have shown promising results in solid tumor therapies. However, identifying functionally specific TCRs is both time-intensive and costly. A new antigen-TCR screening platform, leveraging droplet microfluidics, enables efficient high-throughput screening of pMHC-TCR interactions with heightened sensitivity and minimal background noise. Incorporating DNA barcoding for antigen-presenting and Jurkat reporter cells facilitates specificity assessments of pMHC-TCR interactions, with next-generation sequencing (NGS) offering detailed insights into peptide-MHC-TCR recognition dynamics. This innovative approach promises to streamline the identification of clinically relevant TCRs, enhancing the precision of TCR-T cell therapies.
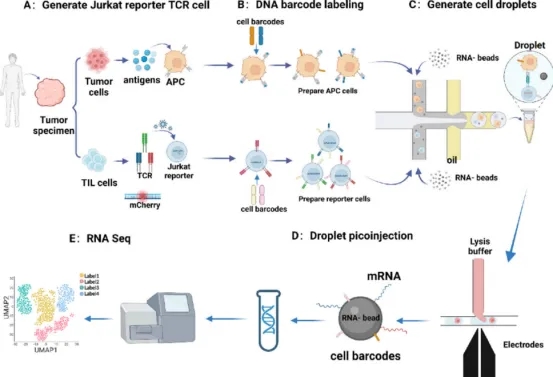
Figure 10. Diagram of the High-throughput Screening Process for Functional TCRs [5]
3. Dominant Neoantigen Validation in Hepatocellular Carcinoma via Co-expression System (Co-HA)
Given the clinical ineffectiveness of many algorithm-predicted neoantigens, empirical validation remains crucial. This study utilized tetramer staining to pinpoint potential neoantigens and introduced a Co-HA for simultaneous expression of patient-specific HLA and antigens. This system facilitated the identification of new dominant neoantigens in hepatocellular carcinoma (HCC) by revealing specific cytotoxic responses against targeted neoantigens, thereby confirming their immunogenicity. Further validation, including tetramer staining and various immunological assays, underscored the potential of these neoantigens in HCC treatment strategies.
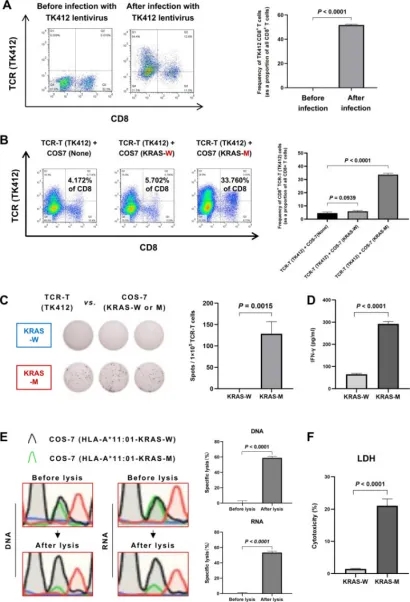
Figure 11. Comprehensive Validation of the Co-HA System [6]
4. Unbiased Identification of CD4+ and CD8+ T Cell Neoantigens via HLA Genetic Screening
Identifying patient-specific neoantigens recognized by T cells poses a significant challenge due to the vast diversity of tumor mutations and HLA alleles. A novel screening method, HANSolo, was developed to detect neoantigens across the full spectrum of a patient’s HLA genotype, employing B cell lines immortalized with Bcl-6/xL and designed to express a wide array of candidate antigens. This unbiased approach allows for the comprehensive identification of T cell-recognized neoantigens, potentially broadening the scope of neoantigen-targeted therapies and enhancing personalized cancer treatment strategies.
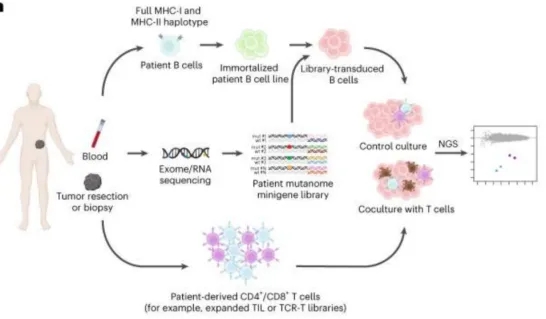
Figure 12. Workflow of the Neoantigen Discovery Process [7]
Sample Submission Requirements
1. Provision of pre-purified samples to the greatest extent possible.
2. Minimization of impurity contamination.
Services at MtoZ Biolabs
1. Complete Experimental Procedures
2. Relevant Instrument Parameters
3. Raw Experimental Data
4. Immunopeptide Screening Analysis Report (including information on antigenic peptide immunogenicity, T cell activation capability, etc.)
Applications
1. Personalized Engineered TCR-T Cell Therapy
Obtaining effective tumor antigen information and accurately sorting TCRs is crucial for designing TCR-T cell therapies for subsequent treatments. The general process includes: Tumor (T) and normal (N) DNA are used for WES and RNA-seq to identify cancer-specific nonsynonymous mutations. Candidate neoantigens are used to design tandem minigene (TMG) encoding mutated peptides and synthesize a mutated peptide library. TILs and PBMCs are isolated from single-cell suspensions obtained from patient samples. Single-cell TCR-CITE-Seq analysis of TILs and PBMCs is performed, using a combination of genetic and surface protein expression tags to predict candidate neoantigen-reactive T cells. Candidate antigen-reactive T cells are co-cultured with autologous APCs expressing candidate neoantigens (TMG or peptides), promoting antigen-specific T cell expansion. Antigen-specific T cells are selected via flow cytometry, and neoantigen-reactive TCRs are identified and screened through scTCR-CITE-seq or deep sequencing. Then, TCRα/β chains are reverse transcribed via single-cell multiplex nested RT-PCR, and their corresponding plasmids are constructed. T cells expressing candidate reactive TCRs are produced by cloning the selected TCR sequences into a retroviral vector and transducing T cells. Recognition of neoantigens by TCRα/β chain-transduced T cells is validated through various screening experiments. PBMCs obtained from the patient are expanded through the aforementioned methods to amplify reactive T cells for neoantigen-specific TCRα/β chains. Verified neoantigen-reactive TCRα/β chains are selected to design the final personalized TCR-engineered T (TCR-T) cell product, which is injected into the patient for cellular therapy.
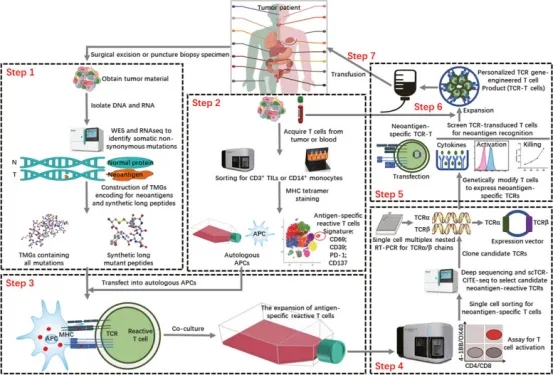
Figure 13. Rapid Identification of Neoantigen-specific TCR for Personalized Engineered TCR-T Cell Therapy [8]
FAQ
Q1: What are the purposes and methods for screening TAAs?
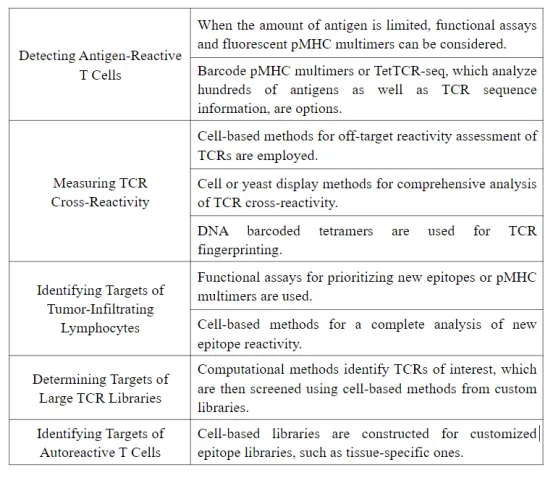
Q2: What are the features of each method?
An article [2] evaluated the features of each method from three aspects: scale and specificity, speed and ease of implementation, and cost and scope of application.
References
[1] Peri, A., Salomon, N., Wolf, Y. et al. The landscape of T cell antigens for cancer immunotherapy. Nat Cancer 4, 937–954 (2023). https://doi.org/10.1038/s43018-023-00588-x.
[2] Joglekar, A.V., Li, G. T cell antigen discovery. Nat Methods 18, 873–880 (2021). https://doi.org/10.1038/s41592-020-0867-z
[3] Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, Haas N, Arlehamn CSL, Sette A, Boyd SD, Scriba TJ, Martinez OM, Davis MM. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017 Jul 6;547(7661):94-98. doi: 10.1038/nature22976. Epub 2017 Jun 21. PMID: 28636589; PMCID: PMC5794212.
[4] Chandran SS, Ma J, Klatt MG, Dündar F, Bandlamudi C, Razavi P, Wen HY, Weigelt B, Zumbo P, Fu SN, Banks LB, Yi F, Vercher E, Etxeberria I, Bestman WD, Da Cruz Paula A, Aricescu IS, Drilon A, Betel D, Scheinberg DA, Baker BM, Klebanoff CA. Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat Med. 2022 May;28(5):946-957. doi: 10.1038/s41591-022-01786-3. Epub 2022 Apr 28. PMID: 35484264; PMCID: PMC9117146.
[5] Li Y, Qi J, Liu Y, Zheng Y, Zhu H, Zang Y, Guan X, Xie S, Zhao H, Fu Y, Xiang H, Zhang W, Chen H, Liu H, Zhao Y, Feng Y, Bu F, Liang Y, Li Y, Xu Q, He Y, Sun L, Liu L, Gu Y, Xu X, Hou Y, Dong X, Liu Y. High-Throughput Screening of Functional Neo-Antigens and Their Specific T-Cell Receptors via the Jurkat Reporter System Combined with Droplet Microfluidics. Anal Chem. 2023 Jun 27;95(25):9697-9705. doi: 10.1021/acs.analchem.3c01754. Epub 2023 Jun 10. PMID: 37300490.
[6] Chen P, Chen D, Bu D, Gao J, Qin W, Deng K, Ren L, She S, Xu W, Yang Y, Xie X, Liao W, Chen H. Dominant neoantigen verification in hepatocellular carcinoma by a single-plasmid system coexpressing patient HLA and antigen. J Immunother Cancer. 2023 Apr;11(4):e006334. doi: 10.1136/jitc-2022-006334. PMID: 37076248; PMCID: PMC10124323.
[7] Cattaneo CM, Battaglia T, Urbanus J, Moravec Z, Voogd R, de Groot R, Hartemink KJ, Haanen JBAG, Voest EE, Schumacher TN, Scheper W. Identification of patient-specific CD4+ and CD8+ T cell neoantigens through HLA-unbiased genetic screens. Nat Biotechnol. 2023 Jun;41(6):783-787. doi: 10.1038/s41587-022-01547-0. Epub 2023 Jan 2. PMID: 36593398; PMCID: PMC10264241.
[8] Li J, Xiao Z, Wang D, Jia L, Nie S, Zeng X, Hu W. The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells. Mol Cancer. 2023 Aug 30;22(1):141. doi: 10.1186/s12943-023-01844-5. PMID: 37649123; PMCID: PMC10466891.
How to order?







