Tumor-Associated Antigens 2 Screening Service
T cells play a vital role in destroying pathogens and monitoring pathological cells. They are able to distinguish between self-antigens and non-self-antigens by surface T cell receptors (TCRs). Most human T cells express the classical TCRs, which consists of TCRα and TCRβ chains co-expressed with CD3 chains (γ, δ, ε, and ζ). The TCRα and TCRβ chains are composed of a variable (V) and a constant (C), respectively. The Vα chain is encoded by V and joining (J) gene segments (TRAV and TRAJ), while the Vβ chain is composed of V, diversity (D), and J gene fragments (TRBV, TRBD, and TRBJ). Three complementarity-determining regions (CDRs) located within the TCR V domain are the main parts of the TCR's interaction with the peptide-MHC (pMHC) complex, thus conferring TCR antigen specificity. Recent results from high-throughput sequencing (HTS) suggest that the actual diversity of the TCR gene pool may be between 1011-1012 unique TCRs. Upon recognition of the homologous pMHC complex by TCR, the immunoreceptor tyrosine-based activation motif (ITAM) in the CD3 chain is immediately phosphorylated, triggering a signaling cascade that leads to T cell activation and induces T cell function.
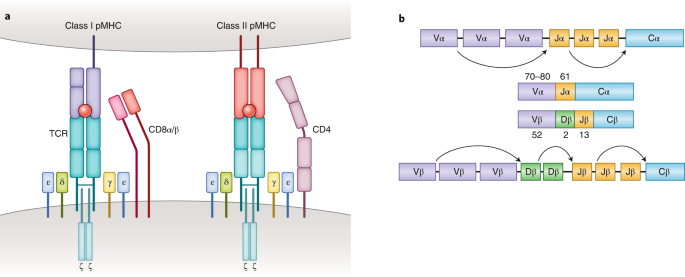
Figure 1. Molecular Basis of TCR-pMHC Recognition[2]
TCR-pMHC interaction is a key factor in adaptive immunity. Classical CD8+ and CD4+ αβΤ cells recognize short peptides presented by the MHC class I and II (MHC I and II) molecules, respectively. In contrast to these MHC-restricted αβ T cells, unconventional γδT cells composed of TCRγ and TCRδ chains are able to recognize non-peptide antigens (e.g., lipid antigens) in an MHC-unrestricted manner. Another characteristic of TCR-pMHC interactions is promiscuity: each TCR is capable of identifying millions of different peptides, and many different TCRs can recognize a given peptide.
The identification and selection of tumor-associated antigens (TAAs) or tumor neoantigens for clinical applications requires more thorough validation rather than simple confirmation as tumor-specific targets. The ideal therapeutic target should have the highest levels of tumor specificity, human leukocyte antigen (HLA) binding capacity, and immunogenicity. Antigens lacking any of these prerequisites, including those that induce T-cell incompetence or tolerance, are not only not therapeutically beneficial, but may even be harmful. Therefore, a series of conditional screening of TAAs and neoantigens is of great significance for the safe development of subsequent treatments.
A variety of antigen-directed methods have been developed to characterize antigen (Ag)-specific T cells. The classic strategy is to detect TCR-pMHC interactions by assessing the functional response of T cells after exposure to a specific antigen. These methods include measuring T cell proliferation, among others. Another commonly used strategy is to use pMHC multimers, which are soluble oligomers of MHC molecules. Four pMHC domains tethered to the central streptavidin molecule by flexible linker sequences and leucine zipper motifs to provide structural stability.
Figure 2. Molecular Design and Application of Class II Tetrameric Molecules[3]
Since it was first reported that pMHC multimers provide adequate TCR binding stability, fluorescently labeled pMHC multimers in combination with flow cytometry have been widely used to visualize and isolate antigen-specific T cells in vitro and in vivo. However, a major factor limiting the scale of T cell response monitoring is the number of different pMHC complexes that can be generated and used simultaneously. To overcome this challenge, peptide exchange techniques (e.g., ultraviolet exchange) have been established to facilitate the rapid generation of various pMHC multimers.
Figure 3. Ag-Specific T Cells Were Identified by Staining with Fluorescently Labeled pMHC Multimers[2]
Another major disadvantage of the pMHC multimer approach is that only a small amount of antigen specificity can be detected in a single biological sample due to the limited number of fluorochromes available. To overcome this obstacle, two parallel methods have been developed to simultaneously detect dozens of Ag-specific T cells using a combination of fluorescently labeled multimers encoded. The first method uses a set of two-color-encoded pMHC multimers to analyze up to 28 antigen specificities in a single sample. The second multivalent coding method utilizes all possible combinations of fluorophores to detect and enrich up to 63 different T cell specificities.
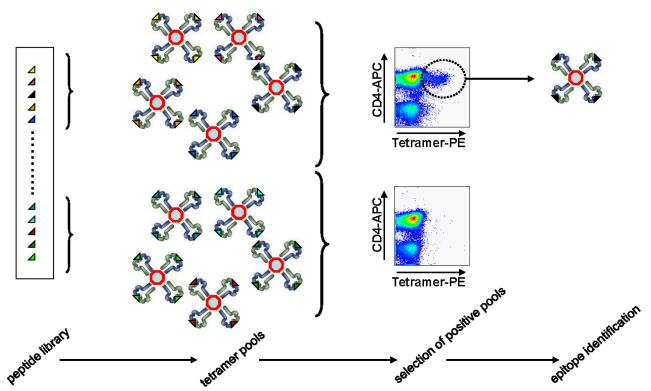
Figure 4. Flow Cytometry Detection of pMHC Multimer Staining to Identify Ag-Specific T Cells [3]
Advances in mass cell measurement techniques using heavy metal ions as labels have provided new strategies to improve the ability of T cell epitope screening. Mass spectrometry (MS) combined with pMHC multimeric staining has resulted in the development of a method to simultaneously identify up to 109 pMHC specificities and an additional 20-30 surface and intracellular phenotypic markers in a single human sample.
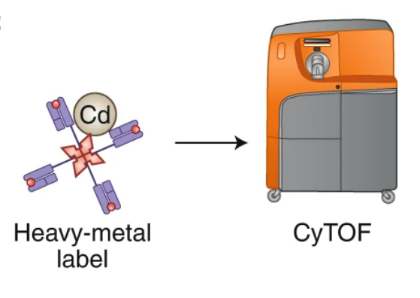
Figure 5. Ag-Specific T Cell Identification by Mass Spectrometry Cytometry (CyTOF)-Based by Heavy Metal-Labeled pMHC Multimers [2]
Recently, this toolkit has been expanded with the use of DNA bar-encoded pMHC multimers. Bentzen et al. have generated DNA-barcoded-pMHC multimers that can simultaneously detect more than 1000 peptide specificities in a single clinical sample. In addition, an upgraded version of DNA barcoding technology, tetramer-associated TCR sequencing (TetTCR-seq), can further link antigen specificity to TCR sequences in single cells at high throughput. The researchers fabricated fluorescent streptavidin-conjugated, DNA bar-encoded pMHC tetramers by in vitro transcription and translation (IVTT) and deciphered the TCRα and TCRβ sequences of individual cells by next-generation sequencing (NGS). In addition to this, recent advances in microfluidic technology have provided new research methods for studying TCR-pMHC interactions at higher throughput.
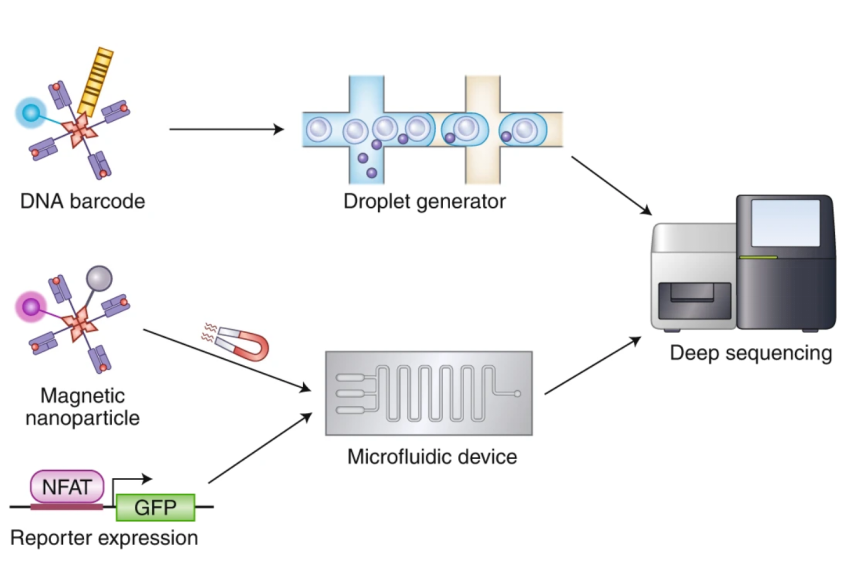
Figure 6. Novel Techniques for Characterizing TCR-pMHC [2]
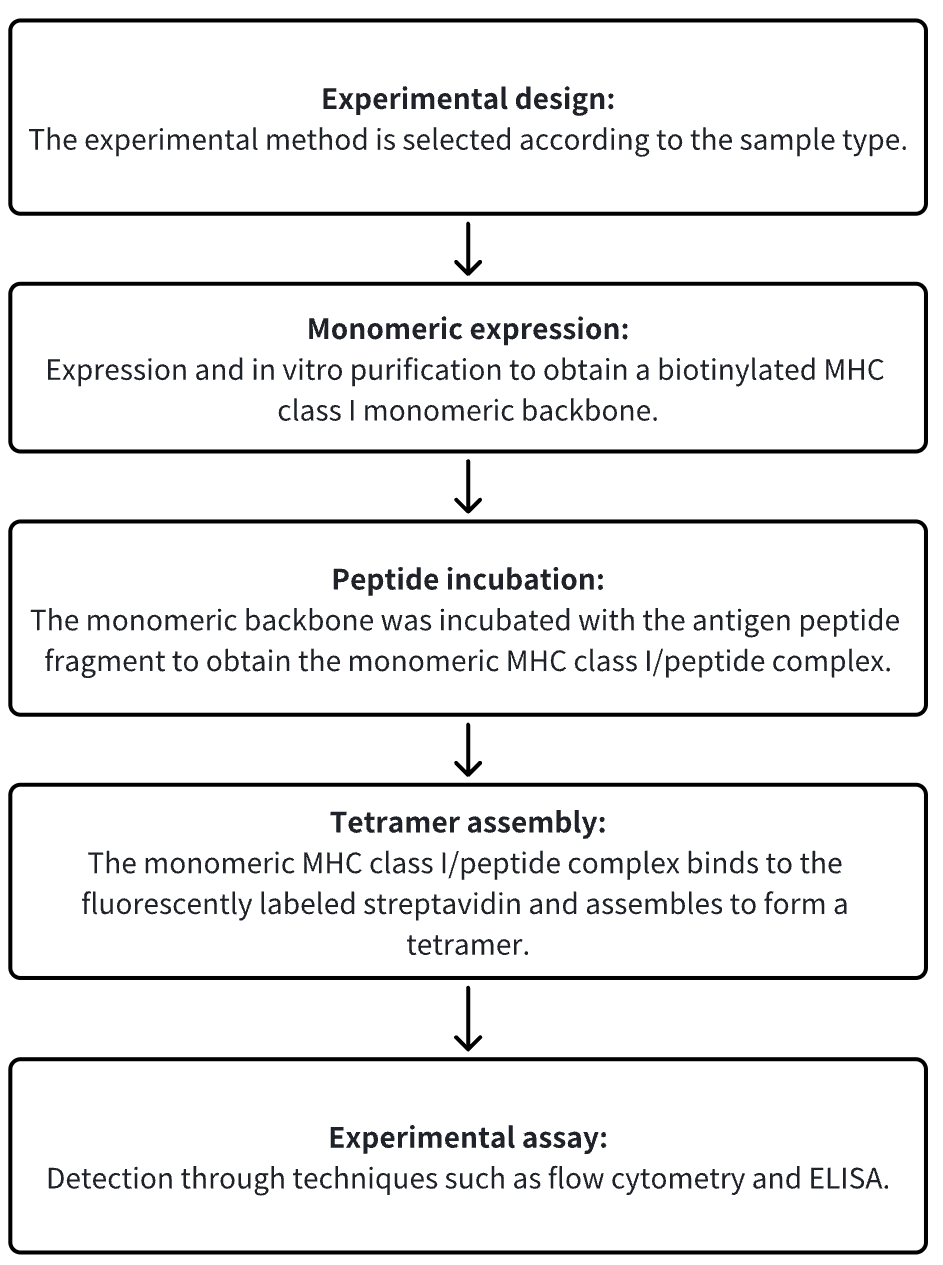
Analysis Workflow
1. Choose the Experimental Method Based on the Specific Requirements of the Experiment
2. Monomeric Expression
3. Peptide Incubation
4. Tetramer Assembly
5. Experimental Assay
Service Advantages
1. Custom development services for natural conformational MHC-peptide complexes.
2. Tetramer products with high purity and high stability (such as tumors (WT1/ gp100/ Mart-1/ p53/ Her-2), viruses, bacteria (CMV/ EBV/ AdV/ HSV/ HHV/ VZV/ Influenza/ HTLV-1/ HIV/ HBV/ HCV/ HPVM.tuberculosis), others (Insulin/ Tetanus Toxin/ OVA/ MOG)).
3. A HTS platform was used to detect the binding effect between MHC-antigen peptide complexes and TCRs.
Sample Results
1. CyTOF &Tetramer screened HCC-infiltrating HBV-specific T cells associated with recurrence-free survival of hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) usually develops following chronic hepatitis B virus (HBV) infection and responds poorly to immune checkpoint blockade. The antigenic specificities of HCC-infiltrating T cells and their relevance to tumor control have been studied. Unexpanded cells from blood, liver, and tumor tissues from 46 HCC patients were stained using a highly multiplexed peptide--MHC tetramer, and 91 different antigen-specific CD8+ T cell populations were detected, which targete HBV, neoantigens, tumor-related, and disease-independent antigens. High-dimensional immunoassay revealed 5 distinct antigen-specific tissue-resident memory T (Trm) cells , two of which were enriched by intratumoral and intrahepatic HBV-specific T cells and manifested as PD-1loTOXlo, while patients with tumor-infiltrating HBV-specific CD8+ Trm cells showed long recurrence-free survival.
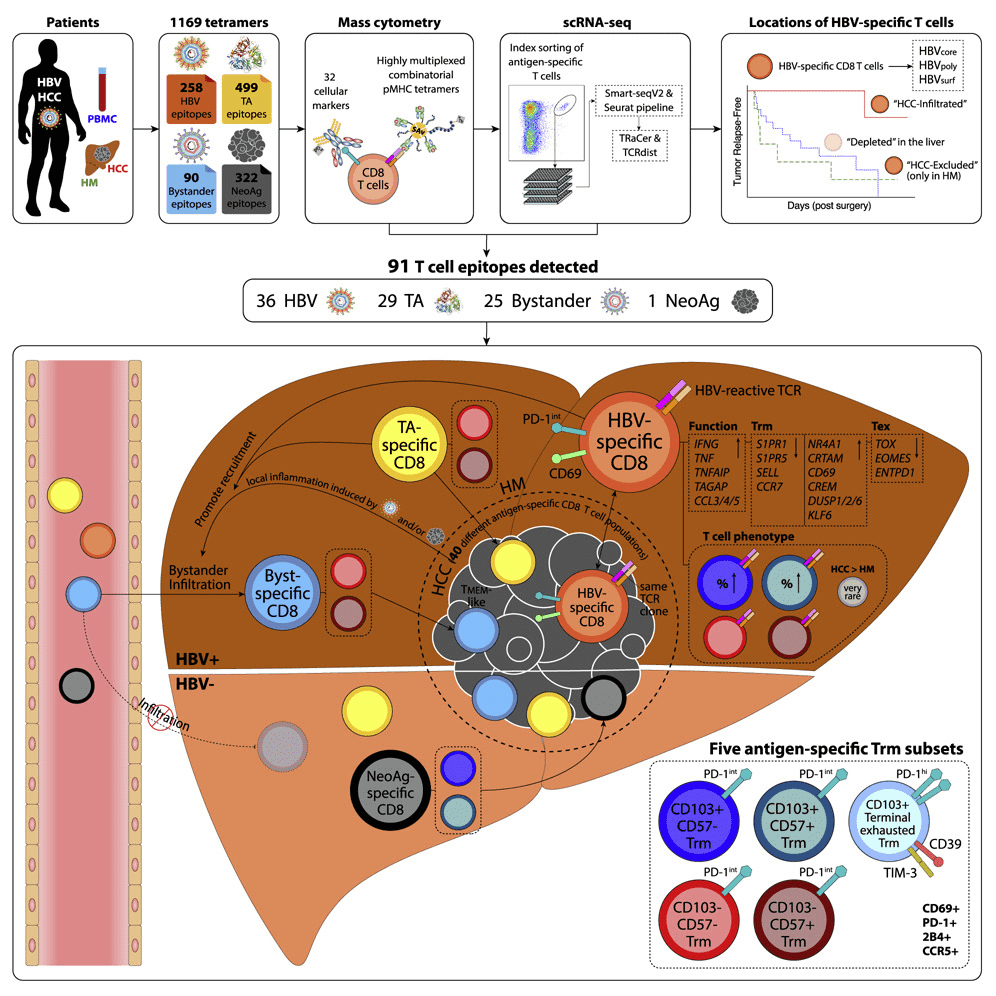
Figure 7. CyTOF & Tetramer Technology Studies the Antigen Specificity of Tumor-Infiltrating T Cells [4]
2. Transplantation elicits a clonally diverse CD8+ T cell response, including potent CD43+ effectors.
CD8+ T cells mediate acute rejection of allografts, which threatens the long-term survival of transplanted organs. Using MHC class I tetramers, allogeneic CD8+ T cells have been found to have a higher naive precursor frequency relative to other epitopes, only a slight increase in the number after grafting, and maintain high diversity of TCRs throughout the immune response. While antigen-specific effector CD8+ T cells poorly express the canonical effector marker KLRG-1, the activated glycoform expression of the activated glycoform CD43 determines potent effector cells after transplantation. Activated CD43+ effector T cells maintain high expression of the coreceptor inducible T cell costimulator (ICOS) in response to CTLA-4 immunoglobulin (Ig), and dual CTLA-4 Ig/anti-ICOS therapy prolongs graft survival. These data suggest that graft-specific CD8+ T cells have a distinct response profile compared to pathogenic CD8+ T cells, with CD43 and ICOS being the critical surface receptors that define potent effector CD8+ T cells population that form after transplantation.
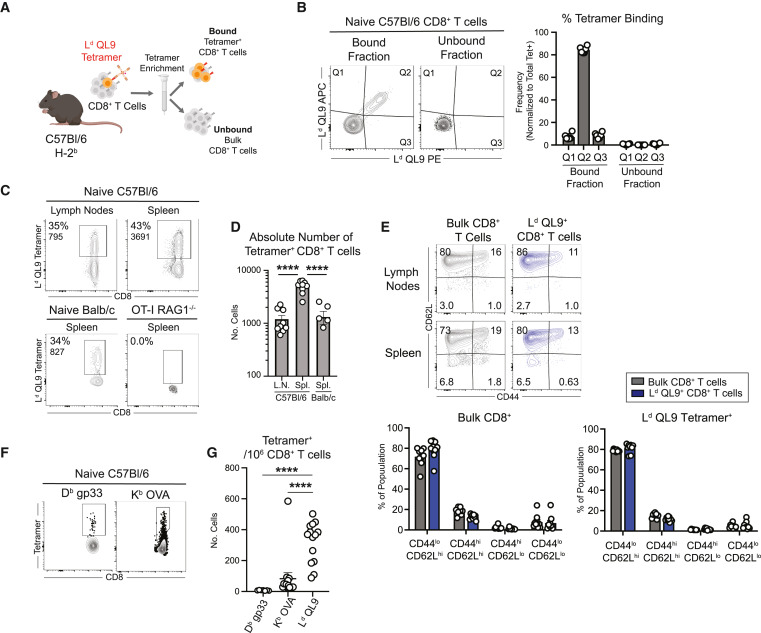
Figure 8. H-2Ld QL9 Tetramers Recognize Naïve CD8+ T Cell Populations with Elevated Precursor Frequencies [5]
3. Large single-stranded trimeric peptide-MHCs libraries enable the discovery and analysis of antigen-specific CD8+ T cells.
The discovery and characterization of antigen-specific CD8+ T cell clonotypes typically involves labor-intensive synthesis and construction of peptide-MHC tetramers. Single-chain trimer (SCT) technology has been applied to high-throughput platforms for pMHC library generation, showing that hundreds of them can be rapidly prepared across multiple class I HLA alleles. Use this platform to explore the effects of peptide and SCT template mutations on protein expression yield, thermal stability, and functionality. SCT libraries are an effective tool for identifying T cells, identifying commonly reported viral epitopes. Then, SCT libraries were constructed to capture SARS-CoV-2-specific CD8+ T cells from COVID-19 participants and healthy donors. The immunogenicity of these epitopes was verified by functional assays of T cells with clonal TCRs captured using SCT libraries. These techniques should enable rapid analysis of peptide-based T cell responses in a variety of situations, including autoimmunity, cancer, or infectious diseases.
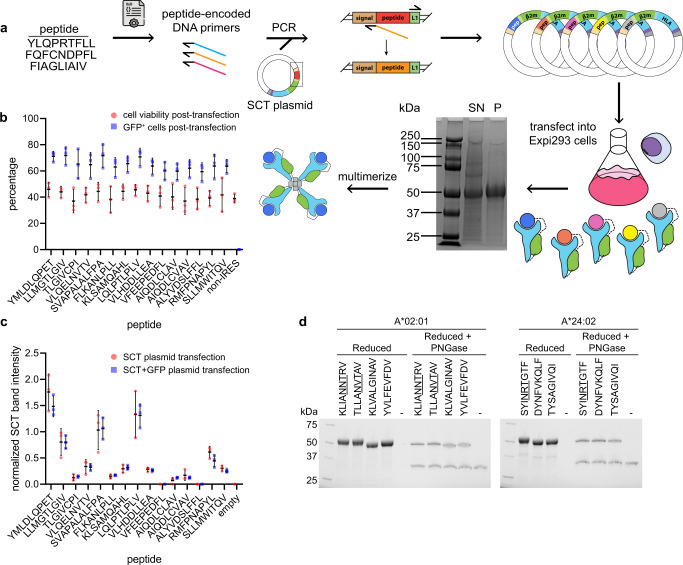
Figure 9. SCT Production and Quality Control [6]
4. Anti-tumor memory CD4+ and CD8+ T cells were quantified by TCR cloning.
A simple, reliable method for detecting anti-tumor memory T cells is necessary for the development of clinical tumor vaccination programs. The clonotype varies in different animals, but is consistent in the blood, spleen, and peritoneal cells (PC) of the same animal. Adoptive metastasis suggests that high-frequency responsive T cells are tumor-specific. Tetramer analysis confirmed that the clonotype frequencies determined by TCR chain-T cell receptor β (TRB) analysis were very close to the cell cloning frequencies. The mean frequency of resting anti-tumor memory CD4+ T cells and memory CD8+ T cells in unchallenged spleens were 0.028% and 0.11%, which was not sufficient to distinguish them from background. Stimulation increased the spleen by an average of ~10-fold and the peritoneal anti-tumor memory T cell clonotype by 100-fold. This method can be used to quickly quantify the effectiveness of tumor vaccines or any vaccine that produces therapeutic T cells using blood and tissue sampling.
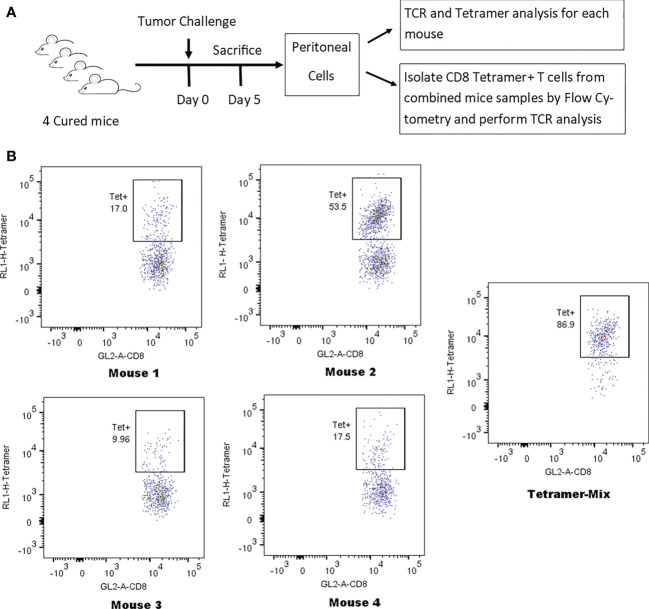
Figure 10. Identification of Tetrameric Clonotypes in Total CD8+ T Cell Populations [7]
Sample Submission Requirements
1. Please provide processed and purified samples whenever feasible.
2. Try to avoid contamination by impurities.
Services at MtoZ Biolabs
1. Complete Experimental Procedures
2. Relevant Instrument Parameters
3. Raw MS Data
4. Immunopeptide Screening Analysis Report (Including Antigen Peptide Immunogenicity, T Cell Activation Ability, etc.)
Applications
1. Ag-specific T cells were detected using in situ MHC tetramer staining.
The development of in situ major histocompatibility complex (MHC) tetramer (IST) staining to detect Ag-specific T cells in tissues has fundamentally revolutionized our understanding of local cellular immune responses to viral and bacterial infections, cancer, and autoimmunity. IST combined with immunohistochemistry (IHC) can determine the location, abundance, and phenotype of T cells, as well as characterize Ag-specific T cells in three-dimensional space relative to adjacent cells and specific tissue locations.
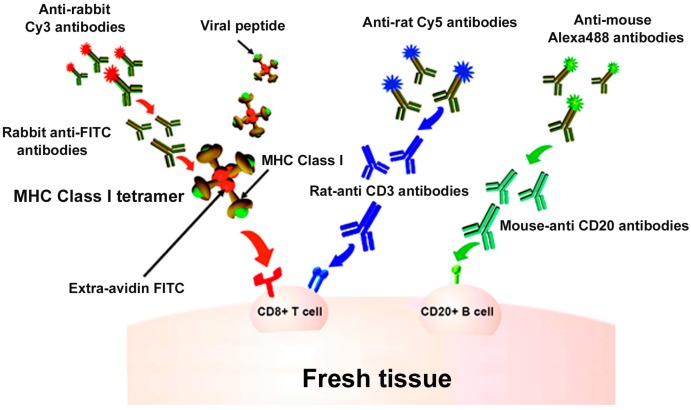
Figure 11. MHCI Tetramer Staining for In Situ Detection of Virus-Specific T Cells [8]
2. HLA class II tetramers are used to determine type 1 diabetes (T1D).
Assessing the risk of developing diabetes, monitoring disease progression, assessing response to treatment, and personalizing antigen-based therapies all require validated assays to measure the frequency and phenotype of autoantigen-specific T cells. To this end, a study has been conducted to verify the tetramer detection of the class II allele HLA-DRA-DRB1*04:01, which is closely related to T1D susceptibility. Immunodominant epitopes of pancreatic islet cell antigens GAD65, IGRP, preproinsulin, and ZnT8, as well as HLA-DRA-DRB1*04:01-restricted T cells specific for reference influenza epitopes, were counted and phenotyped using a tetrameric assay. Single- and multicenter assays were performed using clonal-spike samples and replicate samples from T1D patients with a target coefficient of variation (CV) of less than 30%. The same test was also applied to an exploratory cross-sectional sample set of 24 patients with T1D to evaluate the utility of the test.
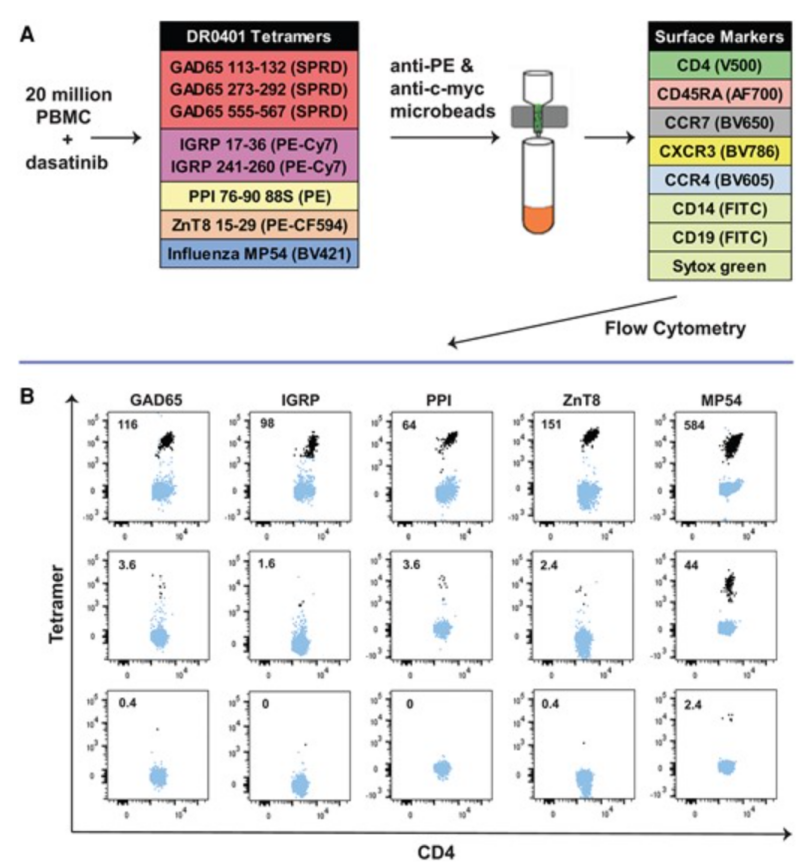
Figure 12. In Vitro Tetramer Staining Process [9]
FAQ
Q1: What are the advantages of tetramer technology?
Compared to other traditional methods used in T cell detection, such as enzyme-linked immunospot (ELISPOT) assays , single-cell PCR, etc., MHC tetramer analysis technology offers the following advantages: High sensitivity: It can detect Ag-specific T cells with low abundance (≤1%) in the blood. High specificity: greatly improves the specificity of TCR binding to MHC-antigen peptide complexes. Good stability: The test results are consistent and reproducible. Diversification/Personalization: Customizable, synthesize-specific antigenic peptide fragments that can be combined into a variety of T cell-selective MHC tetramers. Qualitative/quantitative analysis: Qualitative/quantitative analysis of Ag-specific T cell populations can be achieved in combination with flow cytometry. Flexible choice of fluorescent labels: PE or APC labels are available.
References
[1] Peri, A., Salomon, N., Wolf, Y. et al. The landscape of T cell antigens for cancer immunotherapy. Nat Cancer 4, 937–954 (2023). https://doi.org/10.1038/s43018-023-00588-x.
[2] Joglekar, A.V., Li, G. T cell antigen discovery. Nat Methods 18, 873–880 (2021). https://doi.org/10.1038/s41592-020-0867-z
[3] Nepom GT. MHC class II tetramers. J Immunol. 2012 Mar 15;188(6):2477-82. doi: 10.4049/jimmunol.1102398. PMID: 22389204; PMCID: PMC3297979.
[4] Cheng Y, Gunasegaran B, Singh HD, Dutertre CA, Loh CY, Lim JQ, Crawford JC, Lee HK, Zhang X, Lee B, Becht E, Lim WJ, Yeong J, Chan CY, Chung A, Goh BKP, Chow PKH, Chan JKY, Ginhoux F, Tai D, Chen J, Lim SG, Zhai W, Choo SP, Newell EW. Non-terminally exhausted tumor-resident memory HBV-specific T cell responses correlate with relapse-free survival in hepatocellular carcinoma. Immunity. 2021 Aug 10;54(8):1825-1840.e7. doi: 10.1016/j.immuni.2021.06.013. Epub 2021 Jul 15. PMID: 34270940.
[5] Cohen GS, Kallarakal MA, Jayaraman S, Ibukun FI, Tong KP, Orzolek LD, Larman HB, Krummey SM. Transplantation elicits a clonally diverse CD8+ T cell response that is comprised of potent CD43+ effectors. Cell Rep. 2023 Aug 29;42(8):112993. doi: 10.1016/j.celrep.2023.112993. Epub 2023 Aug 16. PMID: 37590141.
[6] Chour W, Choi J, Xie J, Chaffee ME, Schmitt TM, Finton K, DeLucia DC, Xu AM, Su Y, Chen DG, Zhang R, Yuan D, Hong S, Ng AHC, Butler JZ, Edmark RA, Jones LC, Murray KM, Peng S, Li G, Strong RK, Lee JK, Goldman JD, Greenberg PD, Heath JR. Large libraries of single-chain trimer peptide-MHCs enable antigen-specific CD8+ T cell discovery and analysis. Commun Biol. 2023 May 16;6(1):528. doi: 10.1038/s42003-023-04899-8. PMID: 37193826; PMCID: PMC10186326.
[7] Gao Y, Bergman I. Anti-tumor memory CD4 and CD8 T-cells quantified by bulk T-cell receptor (TCR) clonal analysis. Front Immunol. 2023 Mar 23;14:1137054. doi: 10.3389/fimmu.2023.1137054. PMID: 37033929; PMCID: PMC10076582.
[8] Abdelaal HM, Cartwright EK, Skinner PJ. Detection of Antigen-Specific T Cells Using In Situ MHC Tetramer Staining. Int J Mol Sci. 2019 Oct 18;20(20):5165. doi: 10.3390/ijms20205165. PMID: 31635220; PMCID: PMC6834156.
[9] Ettinger RA, Buitinga M, Vandamme C, Afonso G, Gomez R, Arribas-Layton D, Bissenova S, Speake C, Reijonen H, Kinnunen T, Overbergh L, Mallone R, Kwok WW, James EA. Technical Validation and Utility of an HLA Class II Tetramer Assay for Type 1 Diabetes: A Multicenter Study. J Clin Endocrinol Metab. 2023 Jul 20:dgad434. doi: 10.1210/clinem/dgad434. Epub ahead of print. PMID: 37474341.
How to order?







