Single-Cell Proteomics Analysis
Single-cell proteomics (SCP) is an emerging technology that analyzes the composition, abundance, and modification status of proteins within individual cells, providing revolutionary insights into cellular heterogeneity, biological system complexity, and disease mechanisms. In multicellular organisms, cell-to-cell variability is crucial, and traditional bulk-level proteomics often masks these subtle differences. The advent of single-cell proteomics enables the exploration of unique functions and biological processes at the individual cell level, making it highly applicable to cancer research, immunology, developmental biology, and more.
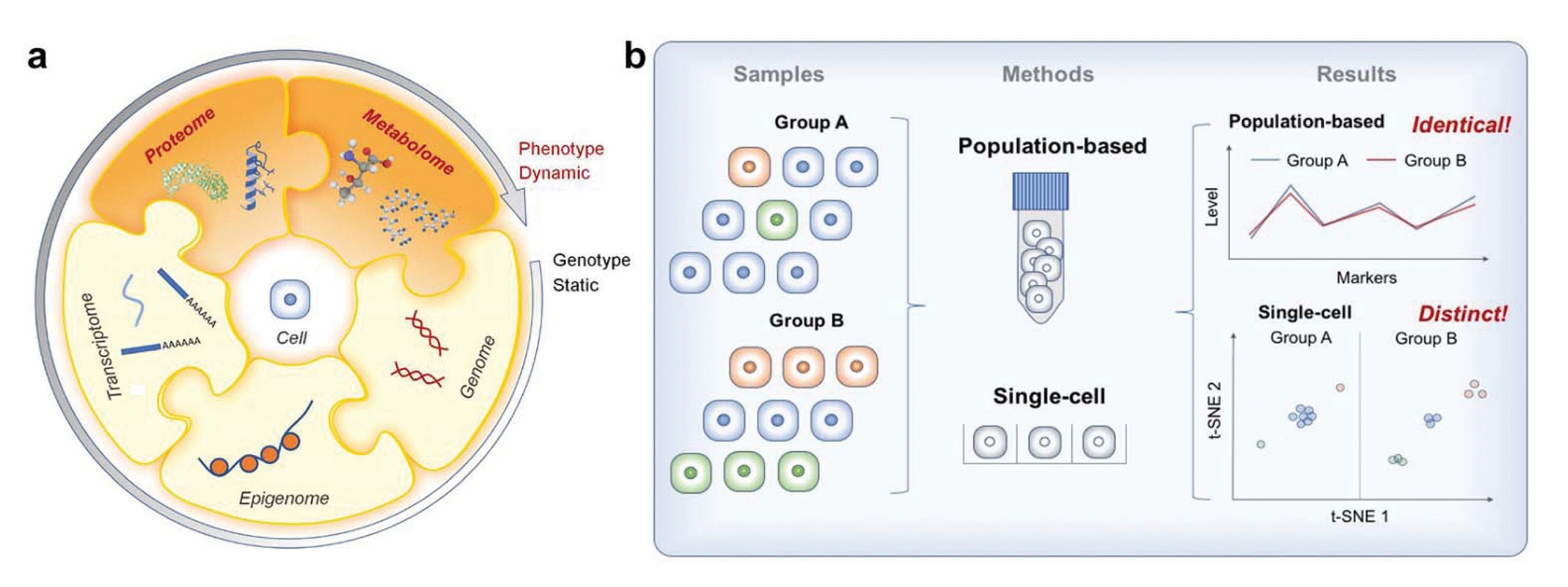
Liu. Y, et al. Analyst. 2019.
Services at MtoZ Biolabs
MtoZ Biolabs is committed to providing cutting-edge single-cell proteomics analysis services. Leveraging advanced mass spectrometry and extensive experimental expertise, we perform protein quantification and modification analysis on nano-scale sample volumes, helping clients uncover biological characteristics at the single-cell level.
Our service addresses critical research challenges: achieving precise single-cell proteomic analysis in the context of high biological sample heterogeneity, revealing specific differences between cells, elucidating disease mechanisms, and understanding complex developmental processes. Our professional team not only provides high-quality data but also assists clients with data analysis and interpretation, enabling reliable scientific conclusions and advancing research projects.
Analysis Workflow
The single-cell proteomics analysis workflow at MtoZ Biolabs involves several key steps:
1. Sample Preparation
Single cells are isolated, lysed, and proteins extracted using advanced micro-separation techniques, ensuring protein integrity.
2. Protein Digestion
Specific enzymes are used to digest proteins into peptides, minimizing sample loss to retain maximum peptide information.
3. Mass Spectrometry Analysis
High-sensitivity instruments (such as LC-MS/MS) are employed for qualitative and quantitative analysis of peptides in single-cell samples.
4. Data Analysis
The mass spectrometry data are interpreted through an AI-powered bioinformatics platform, providing comprehensive insights into protein expression profiles, modification features, and biological significance.
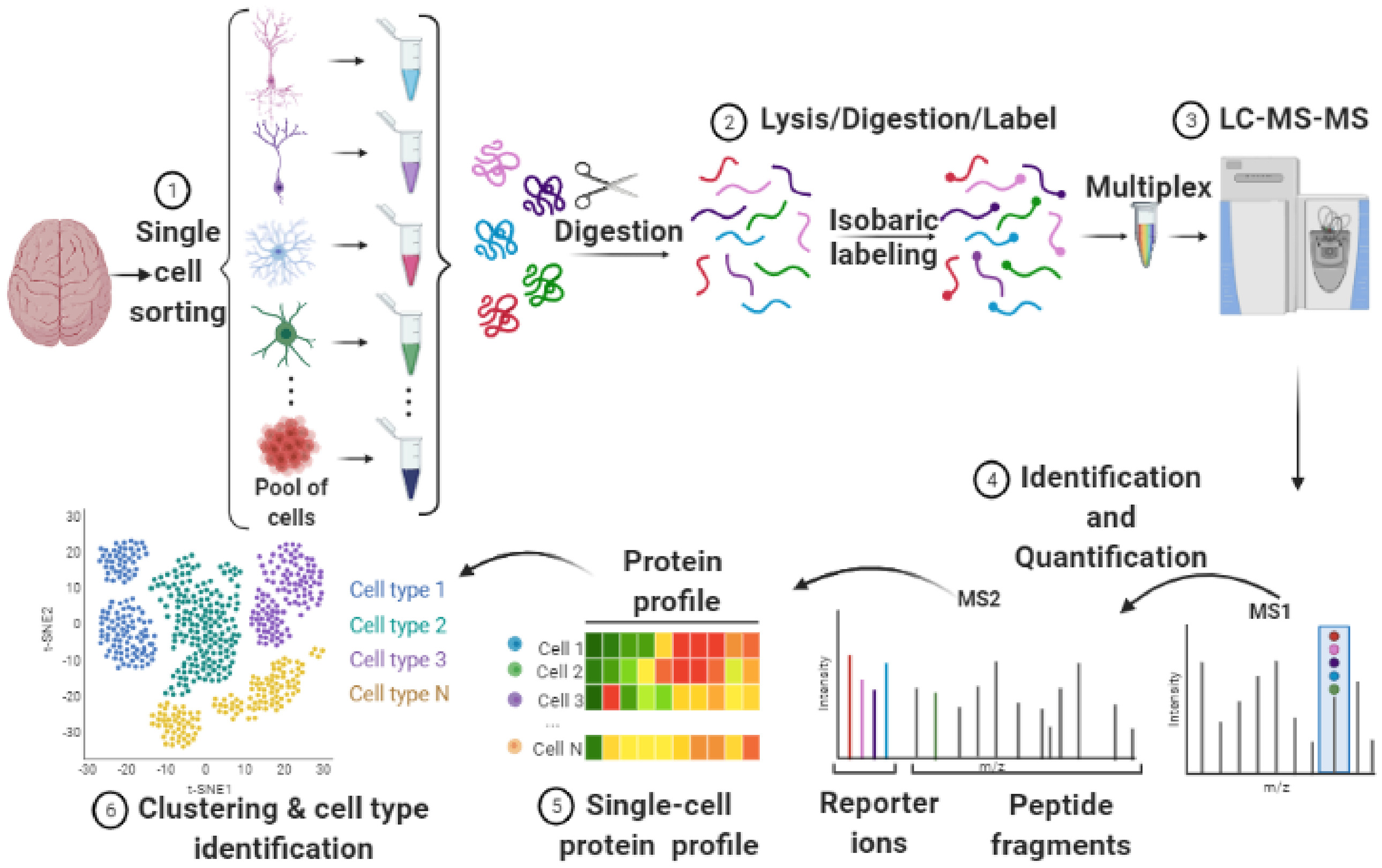
Goto-Silva L, et al. Biochim Biophys Acta Proteins Proteom. 2021.
Pipeline for Single-Cell Proteomics of the Brain
Service Advantages
✅ Advanced Technology Platform for Efficient Analysis of Minute Samples
We utilize industry-leading mass spectrometry platforms, including high-resolution instruments such as the Thermo Scientific Orbitrap Fusion Lumos and Q Exactive. Combined with cutting-edge single-cell proteomics technologies like SCoPE-MS and nanoPOTS, our platform is optimized for the analysis of minute samples. It enables deep coverage and efficient separation of proteins within each cell, ensuring you obtain the most comprehensive single-cell proteomic profiles.
✅ Customized Experimental Design to Meet Diverse Research Needs
Whether your research focuses on specific cell types or requires precise detection of low-abundance proteins, our expert team provides optimal experimental designs tailored to your research goals, including sample preparation, enrichment strategies, and data acquisition. We specialize in utilizing microfluidics for sample separation and offer flexible experimental solutions to accommodate complex research requirements.
✅ Outstanding Quantitative Analysis and Data Processing
With advanced mass spectrometry data acquisition and specialized bioinformatics tools, we achieve high-precision protein quantification at the single-cell level. Our services not only provide comprehensive protein identification but also perform in-depth data mining, assisting you in differential protein expression analysis, functional annotation, and pathway enrichment analysis, thereby supporting a deeper understanding of the complex biological networks within cells.
✅ Strict Quality Control and Experimental Validation
We emphasize accuracy at every step of the experimental process, implementing rigorous quality control measures to ensure data reliability. Additionally, we offer various validation methods, such as Western blotting and immunoprecipitation, to verify key proteins, providing you with greater confidence in your research findings.
Sample Results
1. Pick-up Single-Cell Proteomic Snalysis for Quantifying up to 3000 Proteins in a Mammalian Cell
The shotgun proteomic analysis is currently the most promising single-cell protein sequencing technology, however its identification level of ~1000 proteins per cell is still insufficient for practical applications. Here, we develop a pick-up single-cell proteomic analysis (PiSPA) workflow to achieve a deep identification capable of quantifying up to 3000 protein groups in a mammalian cell using the label-free quantitative method. The PiSPA workflow is specially established for single-cell samples mainly based on a nanoliter-scale microfluidic liquid handling robot, capable of achieving single-cell capture, pretreatment and injection under the pick-up operation strategy. Using this customized workflow with remarkable improvement in protein identification, 2449–3500, 2278–3257 and 1621–2904 protein groups are quantified in single A549 cells (n = 37), HeLa cells (n = 44) and U2OS cells (n = 27) under the DIA (MBR) mode, respectively. Benefiting from the flexible cell picking-up ability, we study HeLa cell migration at the single cell proteome level, demonstrating the potential in practical biological research from single-cell insight.
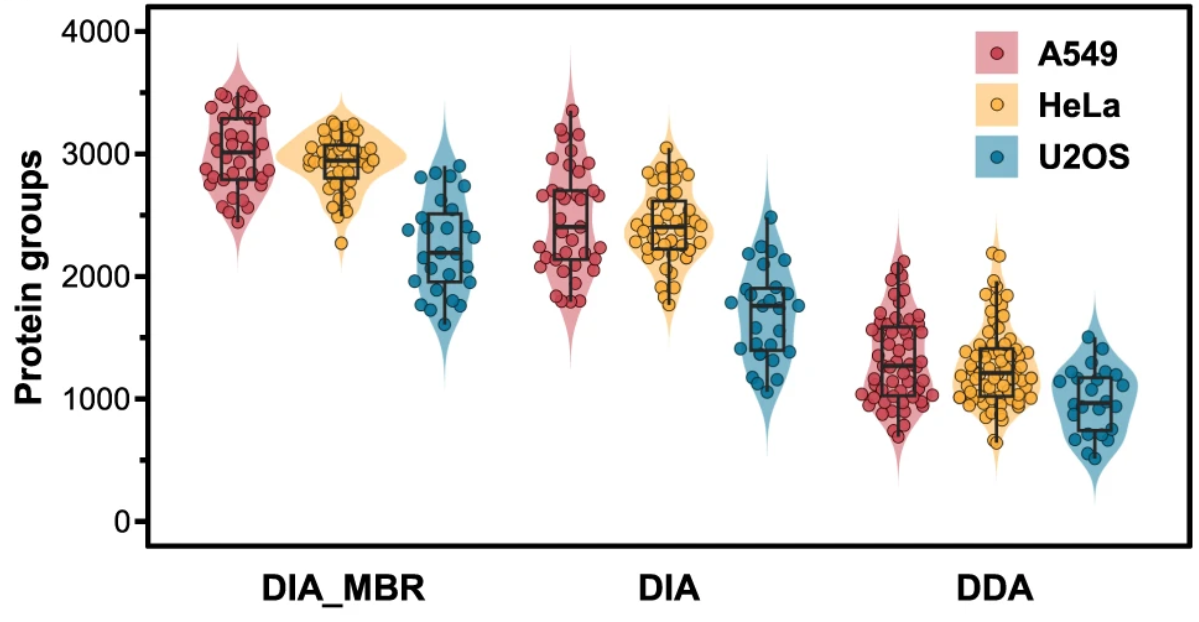
Wang, Y. et al. Nat Commun. 2024.
Quantified Protein Group Numbers of Three Types of Mammalian Cells Under the DIA and DDA Modes
Sample Submission Requirements
1. Sample Type: Single-cell suspension is recommended, and cells should be free of contamination and highly viable.
2. Sample Quantity: At least 500 single cells are recommended for each analysis to ensure accuracy and success rate.
3. Other Requirements: Samples should be free of extraneous proteins, such as serum proteins or cell culture media components.
Our single-cell proteomics service provides strong support for your research, helping you achieve groundbreaking discoveries in both basic research and clinical translation. Please contact our team to learn more about the service or to discuss your specific research needs.
How to order?







