Activity-Based Protein Profiling (ABPP) Service
Activity-based protein profiling (ABPP) is a versatile and powerful chemoproteomic technique, utilizing activity-based probes (ABPs) that selectively and covalently interact with the active sites of proteins. This approach allows for the capture and analysis of tagged proteins using various proteomic tools. ABPP facilitates the global and quantitative examination of protein functional states in complex biological systems, including intact cells and animal models.
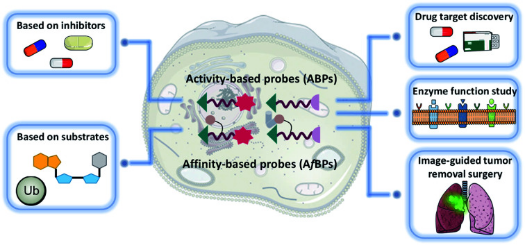
Figure 1. Applications of ABPP [1]
The effectiveness of the ABPP method hinges on the design of the probes, which are small molecules specifically engineered to covalently bind to the active sites of target proteins. Probes are categorized into two types based on their warhead: activity-based chemical probes (ABPs) and affinity-based chemical probes (AfBPs). ABPs are equipped with an electrophilic reactive moiety (warhead) that is specifically tailored to covalently label the catalytically active nucleophilic residues of particular proteins or protein families in a selective and irreversible manner. Typically, an ABP consists of a reactive warhead, a reporter group, and a linker. The warhead, often derived from known covalent inhibitors of the target enzyme, allows for selective marking. Commonly used reporter groups include fluorescent dyes and biotin, which aid in the visualization and enrichment of proteins. The linker, which could be a hydrophilic or lipophilic chain or a peptide, serves to adequately space the warhead from the reporter group.
ABPs contain an electrophilic reactive group designed to irreversibly and selectively label the catalytically active nucleophilic residues of specific proteins or protein families. Alternatively, AfBPs contain a highly selective recognition motif coupled with a photo-affinity group that labels its cognate target protein upon UV-irradiation. The primary distinction between ABPs and AfBPs lies in their selectivity. ABPs react specifically with related classes of enzymes relying on a common mechanism of action, such as the catalytic triad of the serine hydrolases; whereas AfBPs, upon activation, interact with any nearby nucleophilic residue with selectivity for the specific target protein achieved through a classical ligand-protein binding interaction. Thus, while ABP design relies on mechanistic knowledge, AfBPs necessitate prior target knowledge.
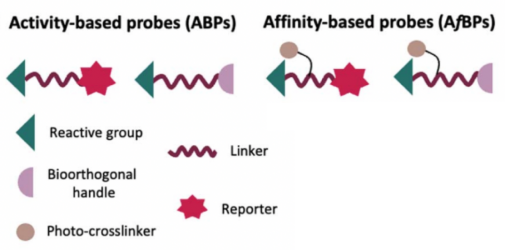
Figure 2. Composition of ABPs and AfBPs [1]
ABPs employ either a one-step or a two-step labeling method. In the one-step method, the probe includes the reporter group, whereas in the two-step method, the reporter unit is added after initial tagging. AfBPs utilize two potential strategies for labelling: a two-step approach involving incubation followed by irradiation, or a three-step method comprising incubation, irradiation, and subsequent bioorthogonal chemistry.
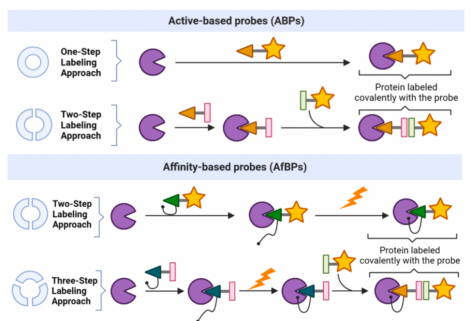
Figure 3. Synthesis of ABPs and AfBPs [2]
The ABPP workflow typically progresses as follows: (1) selection of a target protein; (2) design, chemical synthesis, and characterization of labeling reagents; (3) assessment of labeling efficiency and selectivity under various conditions, such as with purified proteins, cell lysates, or live cells; (4) based on the results of step 3, the process returns to step 2 to optimize the structure of the labeling reagent; (5) application of the optimized probe through methods like visualization, pull-down, and PPI analysis.
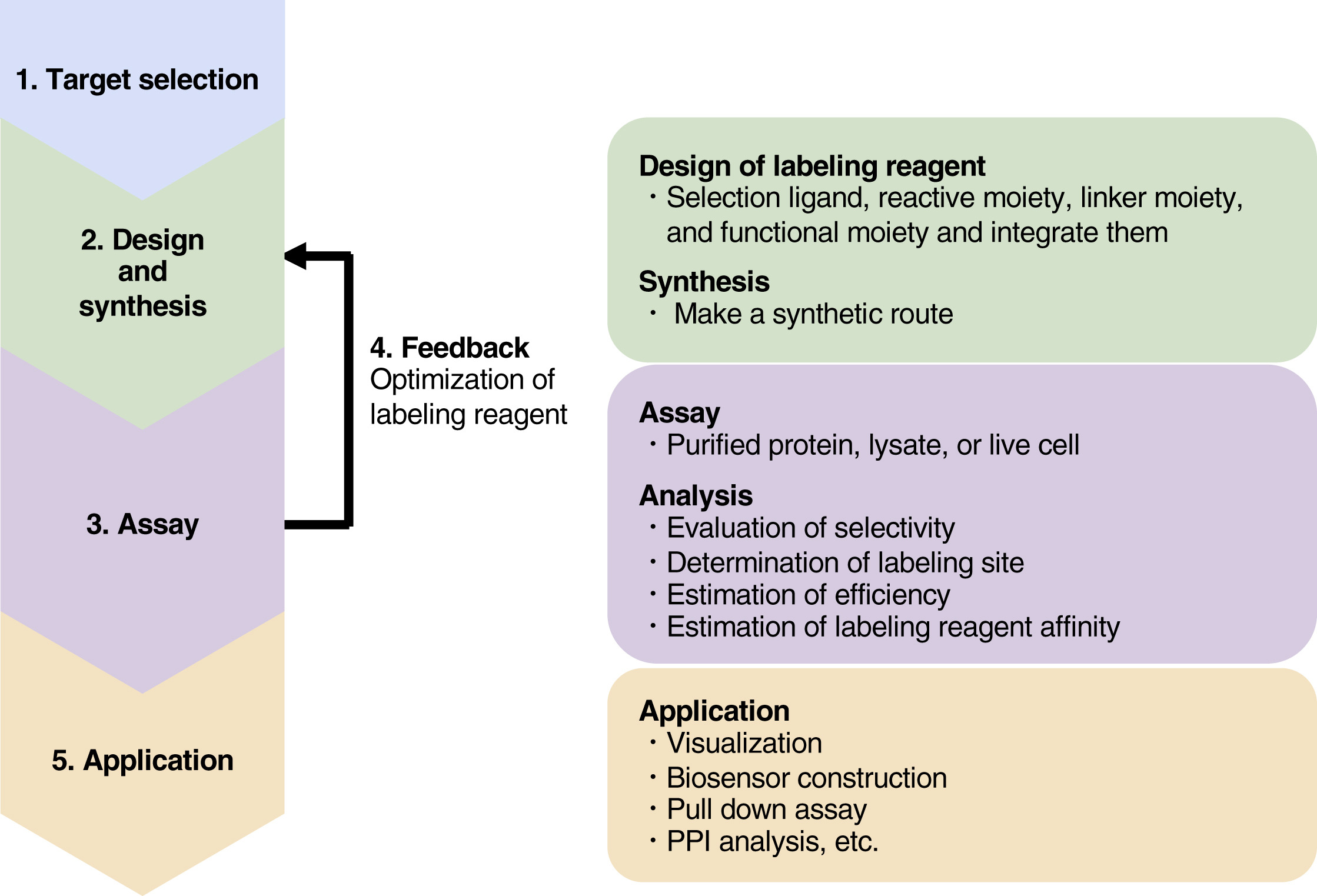
Figure 4. ABPP Workflow [3]
1. Probe Design and Synthesis
(1) Ligand: Choosing the ligand is a crucial step in designing the labeling reagent, as the ligand needs to bind the target protein with high selectivity and affinity. A higher affinity, indicated by a smaller Kd value, generally leads to a faster reaction rate. Detailed information on the structure-activity relationship (SAR) of candidate ligands is beneficial in this step, as the ligand must be derivatized and linked to a reactive molecule or catalyst. Ideally, this linkage should not negatively impact the original properties of the ligand, such as its affinity and selectivity towards the target protein.
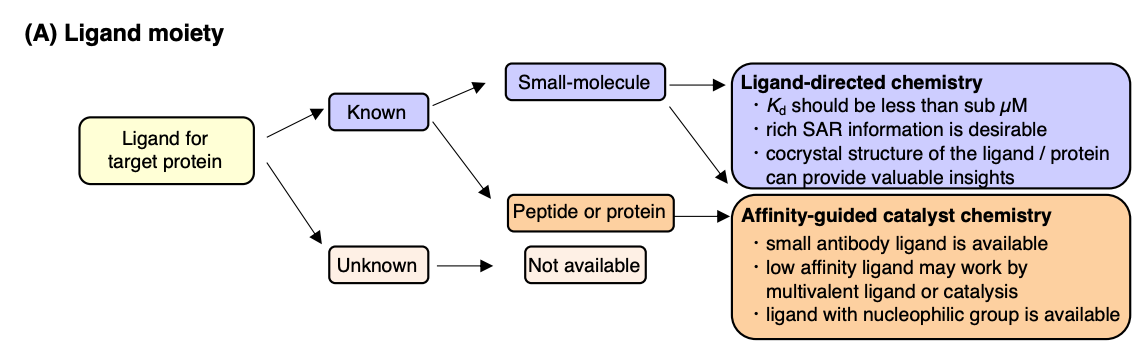
Figure 5. Ligand Selection Strategy [3]
(2) Linker: To maximize the proximity effect for labeling, the linker should be optimized in points of length and rigidity. If a co-crystal structure of the ligand-protein complex is available, the linker length can be designed accordingly. In the absence of a co-crystal structure, it is necessary to prepare labeling reagents with various linker lengths to assess their efficiency and selectivity. For hydrophobic ligands, oligoethylene glycol type linkers are typically used to enhance the hydrophilicity of the labeling reagent.
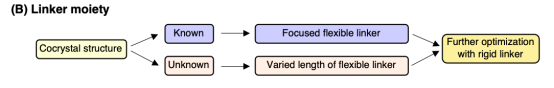
Figure 6. Linker Selection Strategy [3]
(3) Reactive Group: Constructing the reactive part of the probe requires consideration of multiple factors such as the targeted amino acid type, cellular localization, and reaction activity. In live cell labeling experiments, suppressing background reactions with non-target reactive biomolecules is crucial. Researchers have expanded this technique to include additional amino acid types such as methionine, lysine, tyrosine, aspartic acid, and glutamic acid for targeted covalent inhibitor studies.
Table 1. Common Target Amino Acid Reactive Group Structures [4]
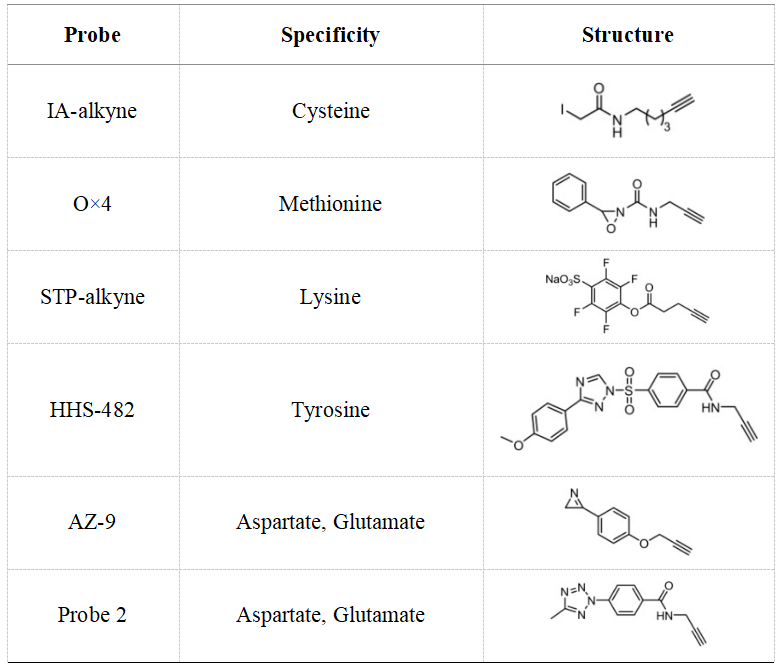
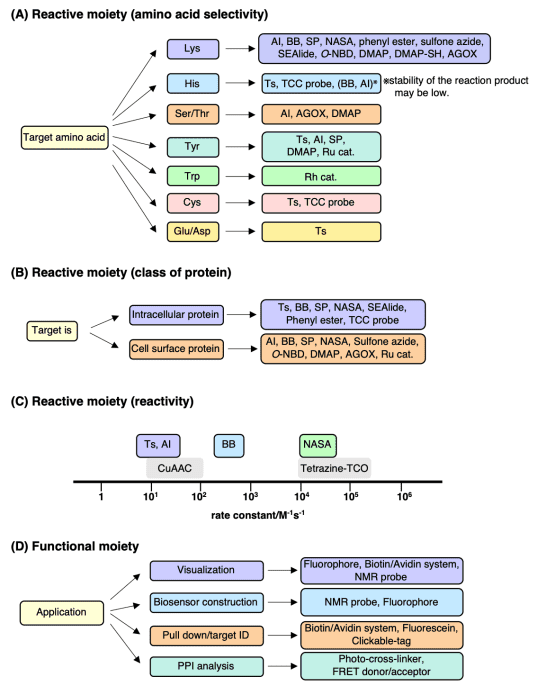
Figure 7. Reactive Group Selection Strategy [3]
2. Qualitative and Quantitative Analysis
ABPP is a technique used to monitor enzyme activities in complex proteomes by integrating chemical probes with quantitative proteomics. Typically, proteomes in cells or in vitro are labeled with ABPs targeting specific enzyme classes or amino acid types. When using fluorescent tags, the labeled proteins can be separated by SDS-PAGE and visualized directly by fluorescence scanning. Alternatively, biotinylated proteins are enriched on streptavidin-immobilized resin and digested to generate probe-modified peptides for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. This allows for the identification or quantification of differences in protein labeling between independent samples, such as healthy and diseased tissues, facilitating the discovery of new drug targets or sites for inhibitor development.
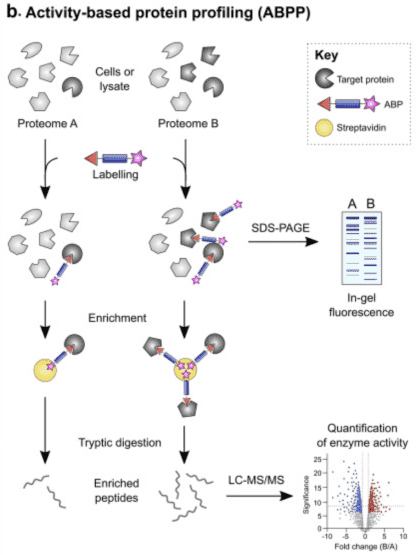
Figure 8. ABPP Experimental Flow [4]
The ABPP analysis process can be adapted into a competitive workflow. Here, proteins are pre-incubated with target ABPs, compounds, or fragments that covalently bind at the same binding site utilized by the chemical proteomics probe. Subsequent to this, the probe labels the remaining sites. After enrichment and washing, the remaining proteins are identified by quantitative proteomics readings. Compared to carrier control or concentration-response experiments, the targets of ABP, compound, or fragment can be identified.
If it is possible to evaluate both covalent and non-covalent targets within the same experiment, more comprehensive data can be gathered. Kinobeads, a chemoproteomic affinity matrix, are primarily used to analyze the target space of protein kinase inhibitors. In a typical competitive Kinobeads experiment, inhibitors are pre-incubated with lysates containing the target proteins, followed by enrichment of the remaining binding sites using Kinobeads and subsequent identification through quantitative mass spectrometry. To differentiate between covalent and non-covalent inhibitors, additional pre-incubation is performed in cells, followed by washing, lysing, and re-equilibration, during which non-covalent interactions are lost. The subsequent Kinobeads pulldown and proteomics analysis then identify covalent interactors as targets. This method allows for the simultaneous analysis of covalent and non-covalent targets.
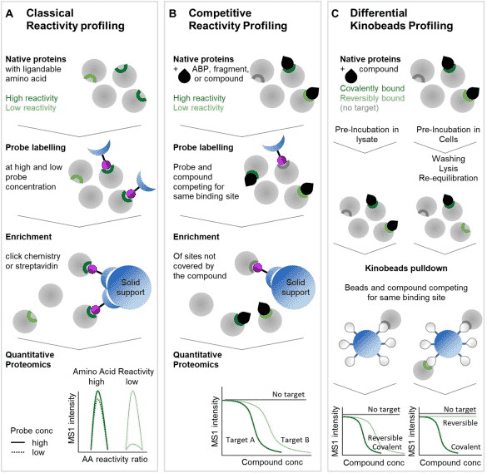
Figure 9. Other ABPP Experimental Flow [5]
Analysis Workflow
1. Experimental Procedure Determination Based on Requirements
2. Probe Design and Preparation
3. Optimization of Probe Design through Multiple Experiments
4. Incubation of Probe with Target Protein/Cells
5. High-Resolution Mass Spectrometry Acquisition Data
6. Data Retrieval and Analysis
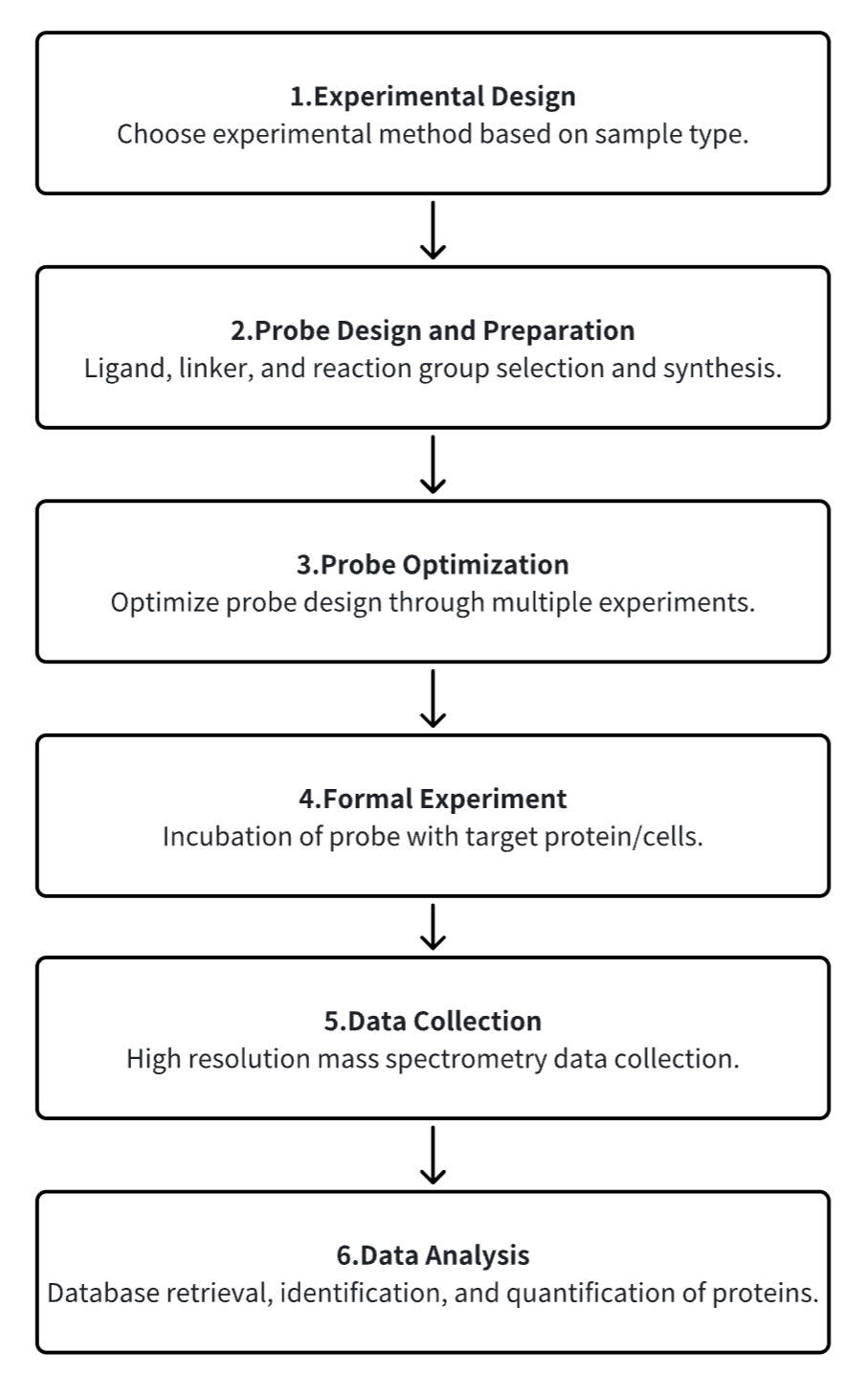
Service Advantages
1. Comprehensive Services for Probe Design, Optimization, and Synthesis
2. High Reliability and Precision in Mass Spectrometry Detection
3. Extensive Bioinformatics Analysis
Sample Results
1. ABPP-CoDEL: Discovery of Tyrosine-Targeting Covalent Inhibitors within a DNA-Encoded Library through Activity-Based Proteome Profiling-Guided
For decades, DNA-encoded chemical libraries (DELs) have been extensively utilized across academia and industry to discover lead compounds. Incorporating an electrophile warhead into DNA-encoded compounds recently permitted the discovery of covalent ligands that selectively react with a particular cysteine residue. However, noncysteine residues remain underexplored as modification sites of covalent DELs. Research has documented the design and utility of tyrosine-targeting DELs of 67 million compounds. Proteome-wide reactivity analysis of tyrosine-reactive sulfonyl fluoride (SF) covalent probes suggested three enzymes (phosphoglycerate mutase 1, glutathione s-transferase 1, and dipeptidyl peptidase 3) as models of tyrosine-targetable proteins. Enrichment with SF-functionalized DELs led to the identification of a series of tyrosine-targeting covalent inhibitors of the model enzymes. Detailed mechanistic studies highlighted their novel action modes and reactive ligand-accessible hotspots of the enzymes. The combined strategy of activity-based proteome profiling with covalent DEL enrichment (ABPP-CoDEL) has yielded selective covalent binders for various proteins, demonstrating this method's potential for further covalent drug discovery.
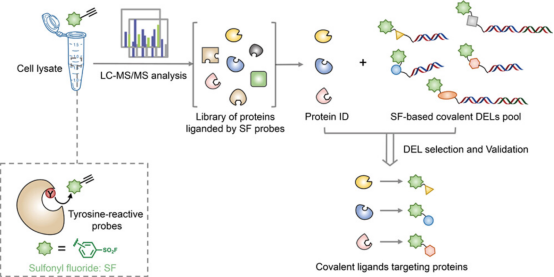
Figure 10. ABPP-CoDEL Experimental Flow [6]
2. Discovery of a Potent Small-Molecule Activity Probe for Broad-Spectrum DUB Analysis in Live Cells
Deubiquitinases (DUBs) form a family of over 100 proteases that hydrolyze the isopeptide bond between ubiquitin and substrates, typically reducing substrate degradation via the ubiquitin-proteasome system. Dysregulation of DUB activity is linked to numerous diseases, such as cancer, neurodegenerative disorders, and autoinflammatory diseases, making them attractive targets for therapeutic intervention. Ubiquitin-derived covalent ABPs are potent tools for analyzing DUB activity spectra; however, their large recognition elements impede cellular permeability, highlighting the need for small-molecule ABPs. These smaller probes could elucidate DUB activity regulation within entire cells or organisms. A comprehensive chemoproteomic warhead analysis led to the identification of the cyanopyrrolidine (CNPy) probe IMP-2373 as a small-molecule pan-DUB ABP. It effectively monitors physiologically relevant DUB activities in live cells. Proteomic and targeted analyses have shown that IMP-2373 quantitatively engages with over 35 DUBs at non-toxic concentrations in various cell lines. Further research has validated its application in pharmacological inhibition and the quantitative analysis of intracellular DUB activity changes during MYC dysregulation in a B-cell lymphoma model, thereby affirming IMP-2373 as a valuable tool for monitoring dynamic DUB activities in disease-related phenotypic backgrounds.
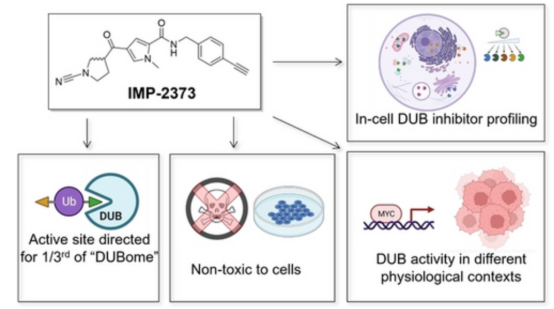
Figure 11. DUB Activity Analysis Using a Small-Molecule Probe [7]
3. Targeted Discovery of Matrine Against PRRSV in Marc-145 Cells via ABPP
Porcine reproductive and respiratory syndrome (PRRS) poses significant challenges to the sustainability of pig industry. Previous studies have demonstrated matrine's effectiveness against the porcine reproductive and respiratory syndrome virus (PRRSV). The research focused on identifying anti-PRRSV targets of matrine in Marc-145 cells employed biotin-labeled matrine 1 and 2 as probes. These probes' anti-PRRSV activities were evaluated through MTT assays, qPCR, and Western blot, and their anti-inflammatory activities were assessed similarly. Using the activity-based proteomics (ABPP) approach, researchers searched for matrine targets in Marc-145 cells and compared these with predictions from network pharmacology to identify potential PRRSV targets. A network database constructed potential target protein-protein interaction (PPI) networks and performed GO/KEGG enrichment analysis. ACAT1, ALB, HMOX1, HSPA8, HSP90AB1, PARP1, and STAT1 were identified as potential matrine targets, linked to its antiviral capacity and immunity.
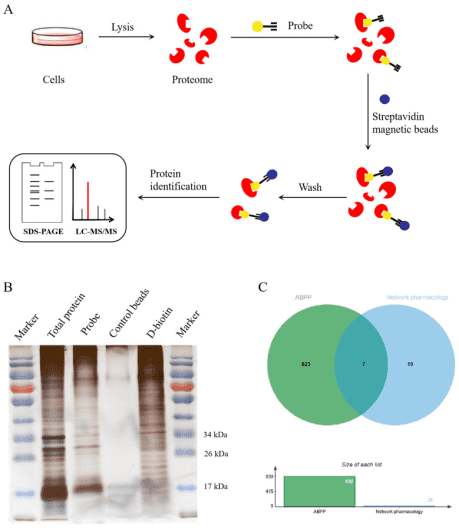
Figure 12. ABPP Experimental Flow [8]
Sample Submission Requirements
1. Samples should not be contaminated with impurities as much as possible.
Services at MtoZ Biolabs
1. Comprehensive Experimental Steps
2. Specifications of Relevant Instrumentation
3. Original Experimental Data
4. Data Analysis Report
Applications
1. Functional Proteomics to Study the Targets and Anticancer Mechanisms of Covalently Acting Natural Products
Eriocalyxin B (EB), 17-hydroxy-jolkinolide B (HJB), parthenolide (PN), xanthatin (XT), and andrographolide (AG) are terpenoids with potent antitumor activities. These compounds typically feature electrophilic groups, such as α,β-unsaturated carbonyls and/or epoxides, enabling them to covalently modify cysteine residues in proteins. Despite their potential, the direct targets and molecular mechanisms of these natural products are not fully understood, which hampers their development. Studies using ABPP combined with quantitative proteomics have methodically characterized the covalent targets of these compounds and their roles in cellular pathways. Initial tests demonstrated the anti-proliferation activities of these five compounds in triple-negative breast cancer cells MDA-MB-231. Quantitative proteomics using tandem mass tags (TMT) showed that all compounds predominantly influence the ubiquitin mediated proteolysis pathway. The ABPP platform pinpointed EB and PN as having significant anti-proliferative activity, with specific targets identified. Biochemical tests indicated that PN inhibits proliferation by targeting the ubiquitin carboxyl-terminal hydrolase 10 (USP10), elucidating a potential molecular mechanism behind its antitumor activity.
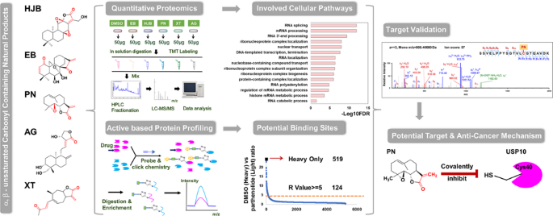
Figure 13. Proteomics Study Flowchart of Targets and Anticancer Mechanisms of Covalently Acting Natural Products [9]
2. Fluorescent ABPs for Cellular Imaging and High-Throughput Screening
Visualization and quantification agents significantly enhance the monitoring of chemotherapy outcomes, early disease diagnosis, and ongoing disease monitoring. Fluorescent ABPs are utilized to visualize and pinpoint enzyme activities within cells or across entire organisms using fluorescence microscopy. These require cell-permeable fluorescent probes or intracellular click-fluorescent reporter genes.
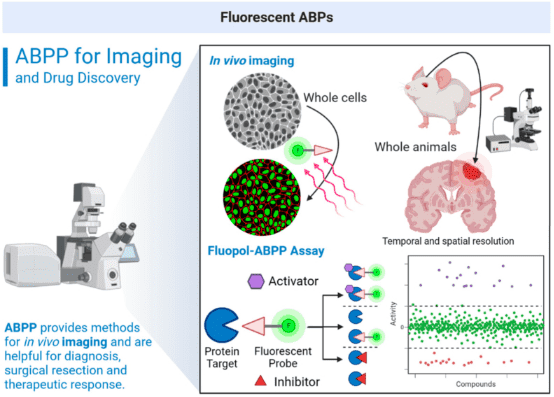
Figure 14. Fluorescent ABPs for Cellular Imaging and High-Throughput Screening [2]
FAQ
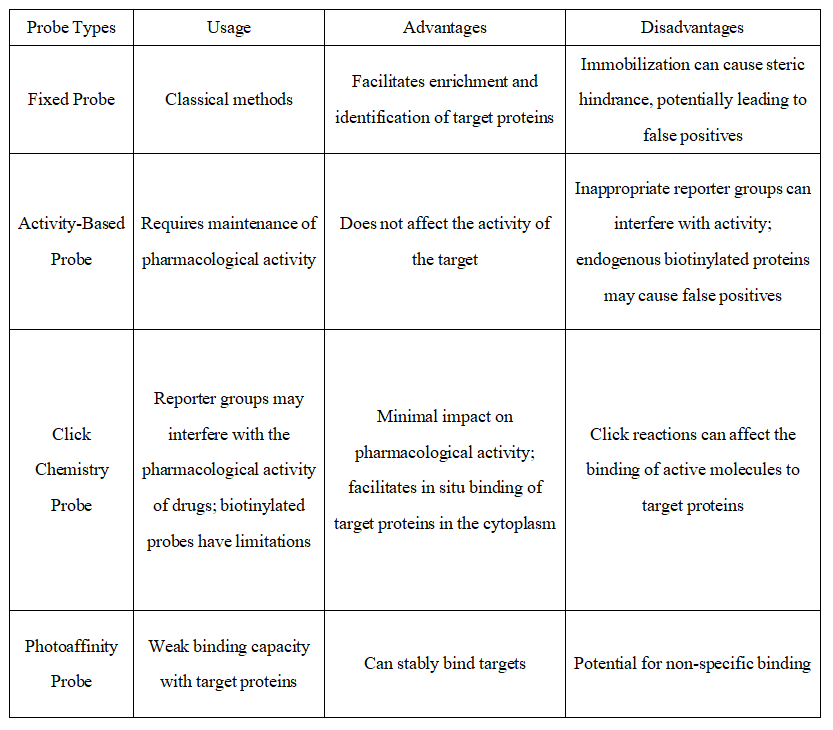
Q1: What are the characteristics of different types of probes?
References
[1] Fang H, Peng B, Ong SY, Wu Q, Li L, Yao SQ. Recent advances in activity-based probes (ABPs) and affinity-based probes (AfBPs) for profiling of enzymes. Chem Sci. 2021 May 18;12(24):8288-8310. doi: 10.1039/d1sc01359a. PMID: 34221311; PMCID: PMC8221178.
[2] Porta EOJ, Steel PG. Activity-based protein profiling: A graphical review. Curr Res Pharmacol Drug Discov. 2023 Aug 24;5:100164. doi: 10.1016/j.crphar.2023.100164. PMID: 37692766; PMCID: PMC10484978.
[3] Shiraiwa K, Cheng R, Nonaka H, Tamura T, Hamachi I. Chemical Tools for Endogenous Protein Labeling and Profiling. Cell Chem Biol. 2020 Aug 20;27(8):970-985. doi: 10.1016/j.chembiol.2020.06.016. Epub 2020 Jul 16. PMID: 32679042.
[4] Benns HJ, Wincott CJ, Tate EW, Child MA. Activity- and reactivity-based proteomics: Recent technological advances and applications in drug discovery. Curr Opin Chem Biol. 2021 Feb;60:20-29. doi: 10.1016/j.cbpa.2020.06.011. Epub 2020 Aug 5. PMID: 32768892.
[5] Heinzlmeir S, Müller S. Selectivity aspects of activity-based (chemical) probes. Drug Discov Today. 2022 Feb;27(2):519-528. doi: 10.1016/j.drudis.2021.10.021. Epub 2021 Oct 30. PMID: 34728376.
[6] Jiang L, Liu S, Jia X, Gong Q, Wen X, Lu W, Yang J, Wu X, Wang X, Suo Y, Li Y, Uesugi M, Qu ZB, Tan M, Lu X, Zhou L. ABPP-CoDEL: Activity-Based Proteome Profiling-Guided Discovery of Tyrosine-Targeting Covalent Inhibitors from DNA-Encoded Libraries. J Am Chem Soc. 2023 Oct 19. doi: 10.1021/jacs.3c08852. Epub ahead of print. PMID: 37857329.
[7] Conole D, Cao F, Am Ende CW, Xue L, Kantesaria S, Kang D, Jin J, Owen D, Lohr L, Schenone M, Majmudar JD, Tate EW. Discovery of a Potent Deubiquitinase (DUB) Small-Molecule Activity-Based Probe Enables Broad Spectrum DUB Activity Profiling in Living Cells. Angew Chem Int Ed Engl. 2023 Oct 1:e202311190. doi: 10.1002/anie.202311190. Epub ahead of print. PMID: 37779326.
[8] Ling X, Cao Z, Sun P, Zhang H, Sun Y, Zhong J, Yin W, Fan K, Zheng X, Li H, Sun N. Target Discovery of Matrine against PRRSV in Marc-145 Cells via Activity-Based Protein Profiling. Int J Mol Sci. 2023 Jul 16;24(14):11526. doi: 10.3390/ijms241411526. PMID: 37511286; PMCID: PMC10381006.
[9] Zhao WS, Chen KF, Liu M, Jia XL, Huang YQ, Hao BB, Hu H, Shen XY, Yu Q, Tan MJ. Investigation of targets and anticancer mechanisms of covalently acting natural products by functional proteomics. Acta Pharmacol Sin. 2023 Aug;44(8):1701-1711. doi: 10.1038/s41401-023-01072-z. Epub 2023 Mar 17. PMID: 36932232; PMCID: PMC10374574.
How to order?







