Advantages, Disadvantages and Principle of Edman Sequencing
Principle of Edman Degradation Sequencing
Edman degradation sequencing, pioneered in the 1950s by Swedish chemist Pehr Edman, is a classic method for determining protein N-terminal sequences. Under mildly alkaline conditions, phenyl isothiocyanate (PITC) selectively reacts with the free α-amino group at the peptide N-terminus, forming a cyclical phenylthiocarbamoyl derivative soluble in organic solvents. Subsequent acid treatment cleaves the terminal amino acid as a detectable anilinothiazolinone (ATZ) derivative, which—after isomerization under acidic conditions—is identified by MS or HPLC.
Iterative cycles “peel” the peptide like an onion, revealing its amino acid sequence residue by residue. Because the chemistry strictly depends on a free α-amino group, the method achieves single-residue resolution with nanomole-scale samples and a theoretical yield of ~99 % per cycle. It remains the gold standard for locating mature protein N-termini, verifying whether post-translational modifications (PTMs) mask the initiator residue, and confirming proteins excised from immunoblots or SDS-PAGE gels—especially for purified peptides or in-gel proteins of 3–30 kDa when database-independent precision is required.
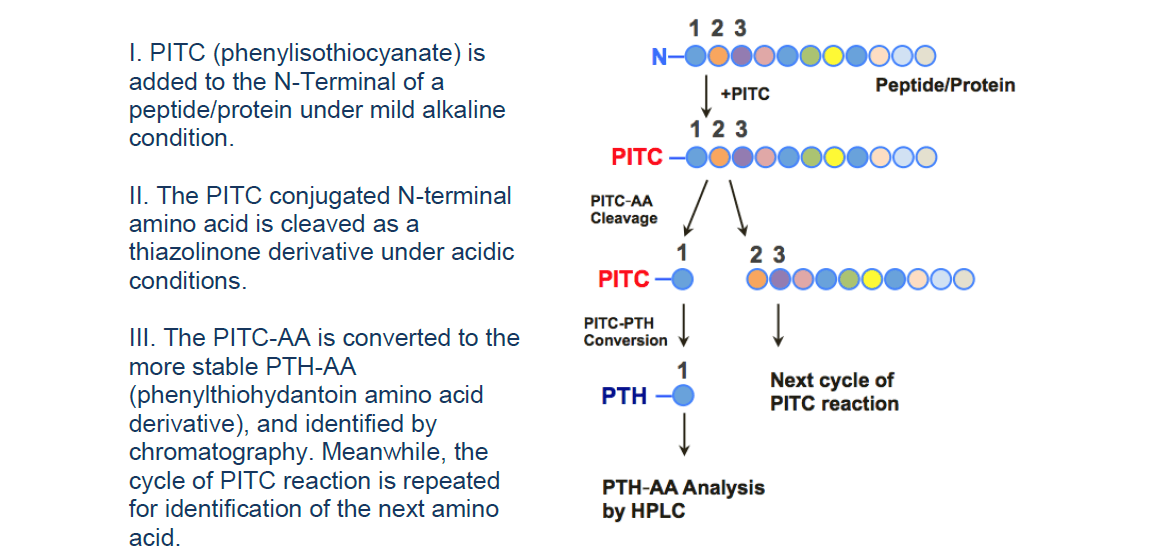
Figure 1. Principle and Workflow of Edman Degradation Sequencing
Advantages of Edman Sequencing
The power of Edman degradation lies in its unrivaled chemical specificity and cycle efficiency. PITC is highly nucleophilic toward free amino groups yet virtually inert to side chains, ensuring a specific reaction site. PTC-peptides dissolve readily in polar organic solvents, eliminating diffusion barriers, while the bicyclic ATZ derivatives possess UV-active conjugated systems that enable rapid, sensitive detection. With solid-phase immobilization (PVDF membranes or glass-fiber filters) and fully automated reactors, modern instruments decode 10–15 residues within 1–2 hours, meeting high-throughput verification needs. In drug development, Edman sequencing is used to determine N-terminal amino acid sequence of antibody light and heavy chains or detect whether recombinant protein drugs undergo partial N-terminal truncation during upstream expression or downstream purification. In mechanistic studies, it pinpoints cleavage sites in inflammatory proteins or signal peptides, facilitating targeted inhibitor design.
Challenges of Edman Sequencing
Yet the method has intrinsic constraints:
(1) Edman sequencing is suitable only for samples whose N-terminal α-amino group is free, without any PTMs;
(2) If a protein contains multiple disulfide bonds, acid-labile modifications, or rare amino acids (such as selenocysteine), acid cleavage can trigger side reactions and reduce Edman sequencing accuracy;
(3) Cumulative derivatization and cleavage losses (~1–2 % per cycle) restrict practical read lengths to 40–60 residues—far shorter than the hundreds accessible by MS-based methods, making Edman sequencing more time-consuming;
(4) Mixed samples or minor N-terminal heterogeneity can obscure overlapping signals;
(5) The need for costly reagents and dedicated automated instruments also inflates per-sample costs for small and medium-scale studies.
How MtoZ Biolabs Addresses the Limitations of Edman Sequencing
MtoZ Biolabs revitalizes Edman sequencing with integrated modern solutions:
(1) Selective deprotection and reduction: enzymatic deacetylation or hydroxylamine treatment unmasks α-amino groups under mild conditions, while DTT/TCEP fully reduces disulfides, boosting initiation efficiency;
(2) Cyclic N-termini are opened chemically or enzymatically before derivatization.
(3) Ultra-micro sample handling: laser microdissection and nano-scale capture columns raise transfer recovery from SDS-PAGE bands by >20 % and lower background noise;
(4) For poorly soluble transmembrane segments, we combine hydrophobic capture agents with reversible surfactants to maintain membrane-protein integrity with high derivatization throughput;
(5) LC-MS validation: high-resolution LC-MS confirms ambiguous UV peaks, preventing residue miscalls;
(6) For sequences >50 residues, the Edman-MS hybrid strategy reads the first 10–15 residues precisely, then extends coverage with high-energy CID/HCD MS, pushing total read lengths beyond 100 residues;
(7) Finally, the in-house algorithm calibrates cycle-efficiency decay, compensates for co-eluting isomers, and generates confidence scores ready for direct integration into LIMS or CMC files.
Conclusion
As the first automated chemical method for protein N-terminal analysis, Edman degradation remains indispensable in proteomics and biopharmaceutical QC. Its absolute chemical specificity, stable cycle efficiency, and independence from sequence databases make it uniquely suited for verifying unknown N-termini, mapping truncations and cleavages, and assessing PTM masking. Edman sequencing is also constrained by drawbacks such as its reliance on a free N-terminal amino group, limited read length, and stringent reaction conditions.
By combining enzymatic deprotection, nano-capture, MS coupling, and intelligent algorithms, MtoZ Biolabs has expanded Edman sequencing’s throughput, accuracy, and read length—providing researchers and drug developers with a flexible, reliable, and cost-effective sequence-verification service. Our upgraded workflow yields rapid N-terminal data from single gel bands at low-nanomole levels and enables segmented validation of complex fusion proteins or membrane fragments, accelerating R&D timelines and deepening structure-function insights.
Contact our technical team for a tailored quotation and discover how MtoZ Biolabs can expedite your next scientific or therapeutic breakthrough.
Related Service
How to order?







