Bottom-Up Proteomics Service
Bottom-up proteomics is a technique that studies proteins by analyzing peptides and is widely used in proteomics research. Its core workflow involves extracting proteins from complex samples, digesting them into peptides using specific enzymes (such as trypsin), separating these peptides through liquid chromatography (LC), and finally detecting and analyzing them using high-resolution mass spectrometry (such as Orbitrap or Q Exactive HF). Bottom-up proteomics enables the identification and quantification of a large number of proteins from complex biological samples and effectively detects post-translational modifications (PTMs). It uncovers protein expression levels, interactions, and functional changes within biological systems, providing critical data for disease research, drug development, and biomarker discovery, making it an essential tool in modern biological and medical research.
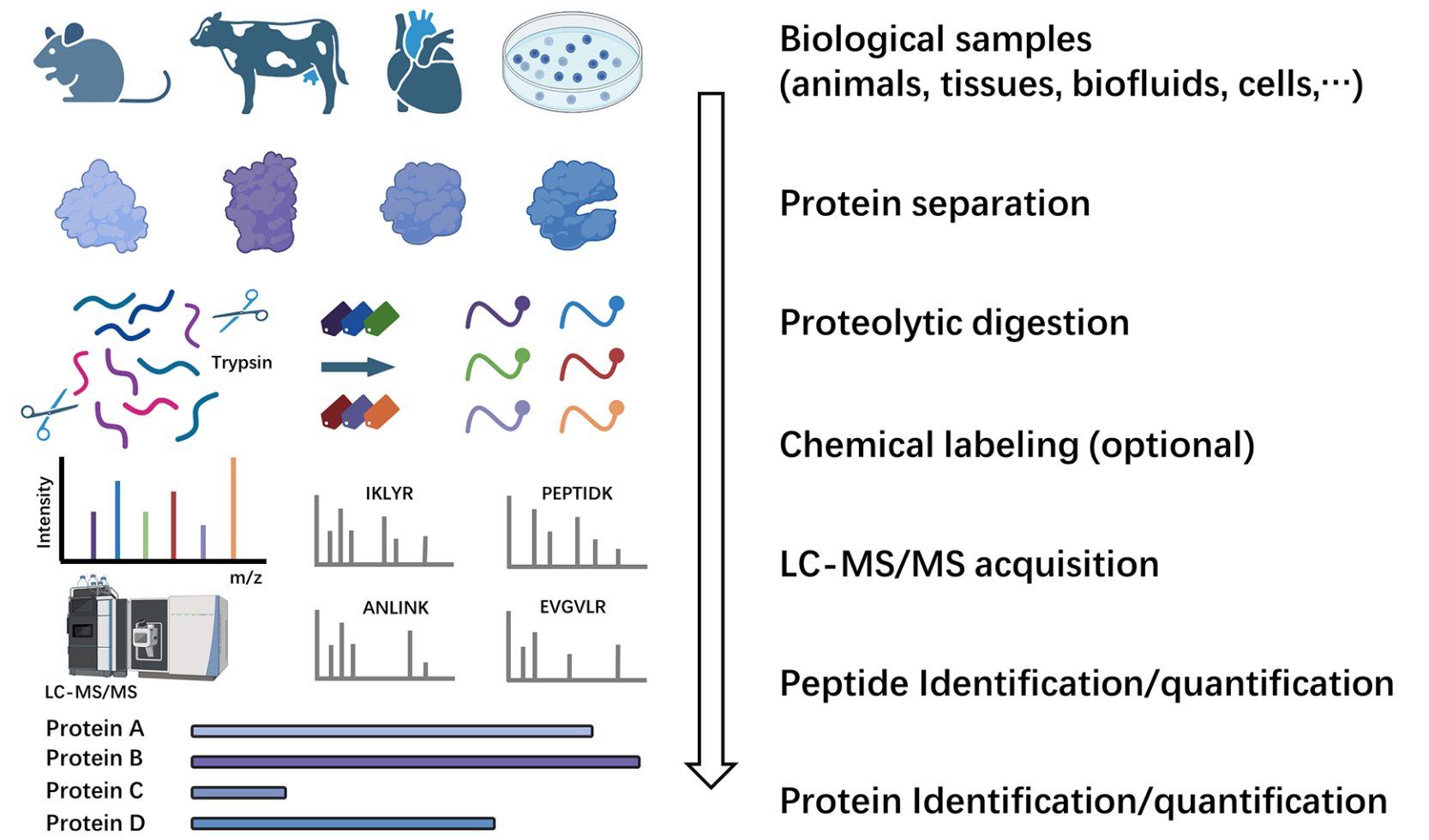
Wang, Z. et al. ACS Meas Sci Au. 2024.
Figure 1. Workflow of Bottom-UP Proteomics
Service at MtoZ Biolabs
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. We have established a powerful and professional bottom-up proteomics platform including Thermo Fisher Q Exactive HF and Orbitrap Fusion Lumos mass analyzer system, coupled with Nano-LC system. Our aim is to provide the most professional support for our clients. If you are interested in our bottom-up proteomics service, please contact us freely.
Service Advantages
1. High-Throughput and High-Sensitivity Analytical Platform
MtoZ Biolabs provides a high-throughput bottom-up proteomics platform suitable for various complex sample types, including clinical samples, large-scale biomarker screening, fundamental scientific research, and multi-omics data integration. Combining high sensitivity with robust performance, the platform ensures rapid analysis of multiple samples while maintaining data quality, effectively addressing challenges such as wide dynamic ranges and the detection of low-abundance biomarkers.
2. Efficient Separation and Optimized LC Technology
MtoZ Biolabs employs optimized multidimensional LC separation techniques for sample fractionation, carefully considering factors such as ionic strength, buffer capacity, and pH effects on retention time and peak shape. This ensures efficient protein or peptide separation. By integrating high-resolution mass spectrometry, the lab achieves enhanced sensitivity and resolution in complex samples, enabling accurate detection of critical low-abundance targets.
3. Tailored Solutions for Diverse Sample Types
Given the challenges posed by the diverse characteristics of proteins, MtoZ Biolabs offers flexible, customized sample preparation and separation workflows. These include techniques ranging from high-abundance protein depletion to affinity-based separations. For each project, the lab optimizes preparation protocols based on sample properties, minimizing protein loss and bias during experimental steps. This ensures reliable and reproducible data, delivering comprehensive analytical solutions for complex samples.
Applications
1. Identification of Disease Biomarker by Bottom-Up Proteomics
Di, Silvestre, Dario. et al. 2016.
2. Bottom-Up Proteomics in Cancer Diagnosis
Ntai, I. et al. Mol Cell Proteomics. 2016.
FAQ
Q1: What are the fundamental differences between Top-down and Bottom-up proteomics methods?
Mass spectrometry (MS) is a key technology in proteomics research. In the field of protein mass spectrometry, two main analytical strategies are commonly used: Top-down and Bottom-up. These two methods differ fundamentally in how they handle and analyze protein samples.
1. Top-down Proteomics
In the Top-down approach, intact proteins are directly introduced into the mass spectrometer and fragmented into smaller pieces within the instrument. This means the analysis starts from the entire protein molecule. The Top-down method enables the direct analysis of a protein’s primary structure, post-translational modifications (PTMs), and protein isoforms. The advantage of this strategy is its ability to accurately characterize the composition and structure of proteins, as it retains the original information of the protein molecule. However, its limitations include the need for high-resolution and high-sensitivity mass spectrometry, as well as stringent sample preparation requirements.
2. Bottom-up Proteomics
In contrast to the top-down method, the bottom-up approach begins with digesting proteins into peptides using specific enzymes, typically trypsin. These peptides are then separated and introduced into the mass spectrometer for analysis. This means the analysis starts from protein fragments. This strategy generally achieves high protein identification rates and coverage even under conditions of lower mass spectrometry resolution and sensitivity. However, since the Bottom-up method loses information about the intact protein structure, it has weaker capabilities in analyzing PTMs and protein isoforms.
In essence, the top-down and bottom-up proteomics differ fundamentally in protein mass spectrometry analysis. The top-down method directly analyzes the intact protein molecule, while the bottom-up method works by analyzing protein-derived peptides. Each approach has its strengths and weaknesses, and researchers must select the appropriate strategy based on their specific goals and requirements.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Liquid Chromatography and Mass Spectrometry Parameters
4. The Detailed Information of Bottom-Up Proteomics Analysis
5. Mass Spectrometry Image
6. Raw Data
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







