Decoding Plant Roots with Single-Cell Precision: Single-Cell Proteomics Reveals Arabidopsis Cell Functions
Single-cell proteomics (SCP) is a cutting-edge analytical technique that deciphers protein composition and function at the resolution of individual cells. By uncovering cellular-level details, single-cell proteomics reveals intercellular heterogeneity and addresses limitations of traditional bulk proteomics, which provide only average or aggregate protein expression data. In plant research, single-cell proteomics circumvents challenges posed by rigid cell walls that hinder traditional methods from accessing individual plant cell proteomes. It enables researchers to examine cellular heterogeneity, unravel gene regulatory networks, and explore cell-specific functions in plants. Leveraging these capabilities, the Iowa State University research team applied single-cell proteomics to Arabidopsis roots, successfully distinguishing endodermal and cortical cells. This achievement lays a theoretical and technical foundation for single-cell studies in plants. MtoZ Biolabs employs the fully automated CellenONE platform to deliver precise single-cell proteomics analyses, offering innovative tools for tackling complex cellular heterogeneity in plant research.
Arabidopsis, a widely used model organism, is ideal for studying cellular heterogeneity due to its well-defined root cell types and spatial organization. As essential organs for nutrient absorption and transport, roots exhibit diverse cell types with unique physiological functions and adaptive responses. Traditional proteomics approaches have inherent limitations in resolving protein expression differences between distinct cell types within root tissues. In contrast, single-cell proteomics offers a groundbreaking solution to this challenge. Through single-cell proteomics analysis of Arabidopsis root cell types, researchers at Iowa State University have provided foundational insights into the functional specialization of root systems and their roles in environmental adaptation. The key findings of their study are summarized as follows:
1. Methodological Advancements and Data Quality Evaluation
Arabidopsis roots, with their clearly defined cell types, serve as a robust system for benchmarking plant SCP. The study focused on two adjacent cell types, cortical and endodermal cells, derived from a shared stem cell lineage (Fig. 1). Protoplasts were enzymatically isolated from roots and sorted into these two cell types using flow cytometry. A total of 384 protoplasts for each cell type were deposited into 384-well plates.
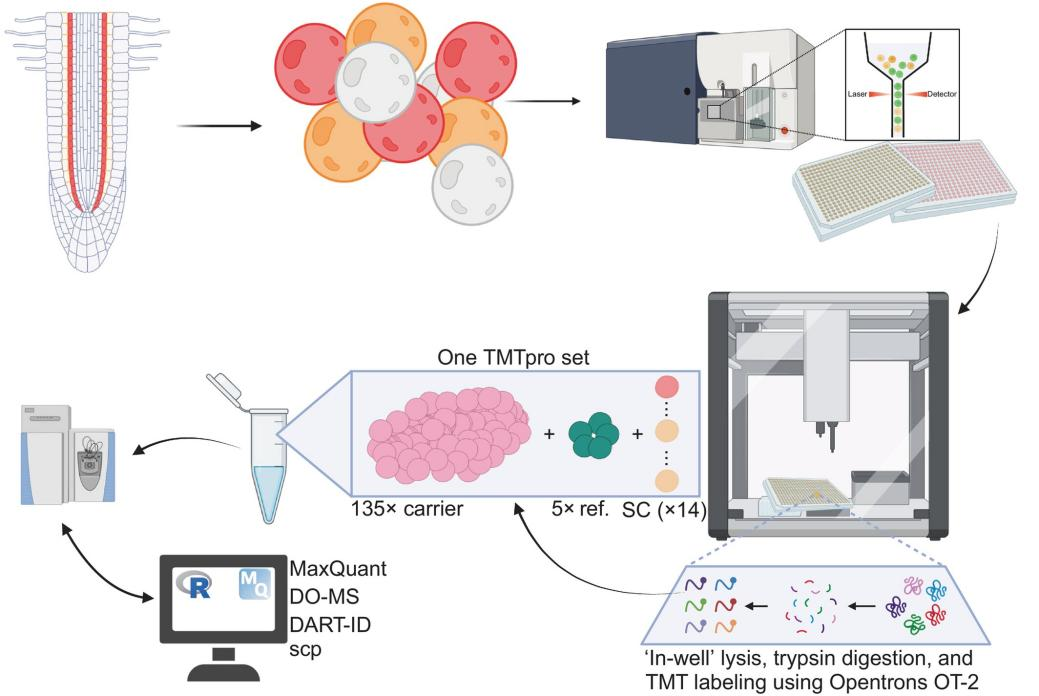
Figure 1. Single-Cell Proteomics Workflow for Arabidopsis Endodermal and Cortical Cells
Optimized protocols for sample preparation and labeling enabled the analysis of 756 individual cells. Raw data, processed using MaxQuant, demonstrated high sensitivity and consistency (Fig. 2). The analysis identified 3,763 proteins, with an average of 1,118 proteins quantified per cell.
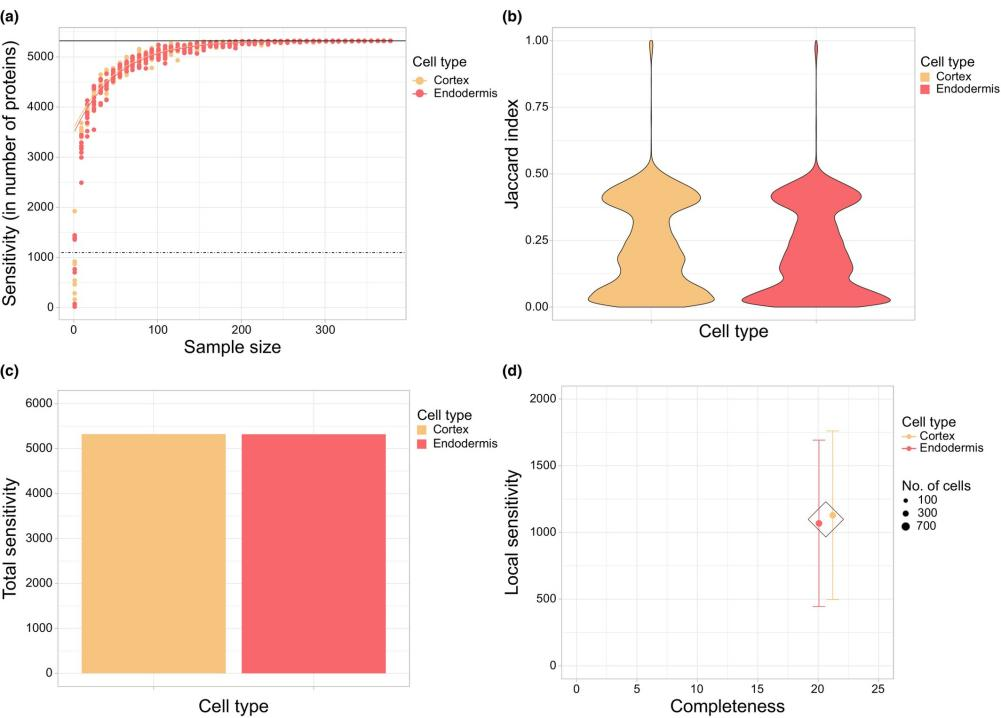
Figure 2. Data Quality Evaluation in Single-Cell Proteomics
Peptide coefficients of variation (CVs) were calculated, and cells with extreme peptide CVs (> 0.69) were excluded. Proteins quantified in fewer than 98% of cells were also removed, resulting in a rigorously filtered dataset. Despite reducing the number of cells to 81 (33 endodermal cells and 48 cortical cells), this stringent filtering still allowed the quantification of 3,217 proteins.This outcome is hypothesized to result from differences in root cell size (total protein per cell), as root cells exhibit a size gradient along the root's length. Additionally, genome duplication events in many plant tissues, including roots, often lead to increased cell size. However, this increase is nonlinear, emphasizing the need to identify normalization factors beyond histones in plant SCP studies. Future efforts to address quantification variability arising from cell size heterogeneity could leverage instruments like CellenONE, which facilitate sorting based on both fluorescence and cell size/morphology.
2. Data Inspection
Quantitative analysis of the identified proteins revealed that SCP detects proteins across a broad range of transcript abundances, with a preference for highly expressed transcripts. Additionally, SCP demonstrated its ability to quantify signaling proteins, often present at low abundance. Among the 3,217 quantified proteins, 175 kinases and 188 transcription factors were detected, highlighting SCP’s capability to capture key signaling molecules.
3. Results Analysis
The proteomic differences between endodermal and cortical cells were analyzed using the rigorously filtered dataset. Principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) analyses of the top 25% most variable proteins revealed clear separation between endodermal and cortical cells (Fig. 3).
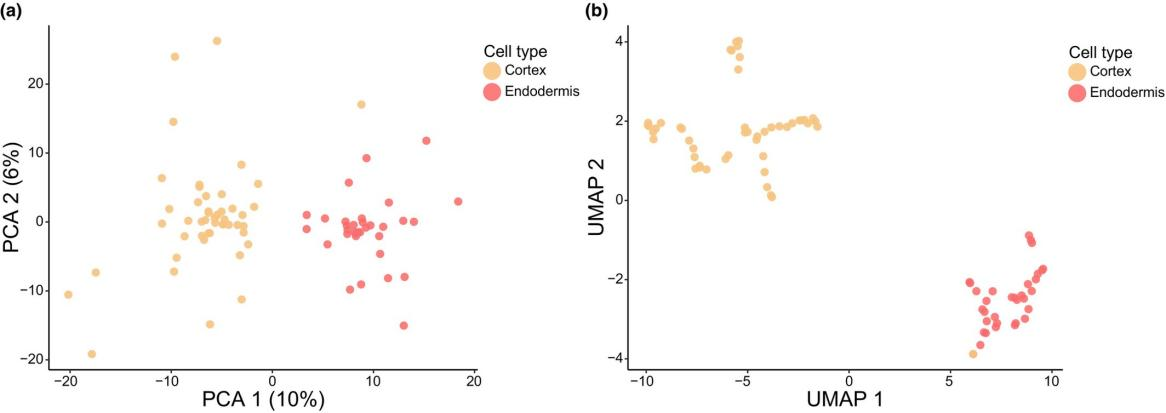
Figure 3. PCA and UMAP Analysis of Proteome Variability
Differential expression analysis identified 596 proteins with significant differences between the two cell types (Fig. 4a). Enrichment of cell-type-specific markers validated the results. For instance, CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN 1, essential for Casparian strip formation, was enriched in endodermal cells, along with RESPIRATORY BURST OXIDASE HOMOLOG D, known to activate reactive oxygen species (ROS)-dependent signaling. Similarly, NAKED PINS in YUCCA 2 was enriched in endodermal cells, consistent with its role in regulating PIN-FORMED 1 localization and gravitropic responses.
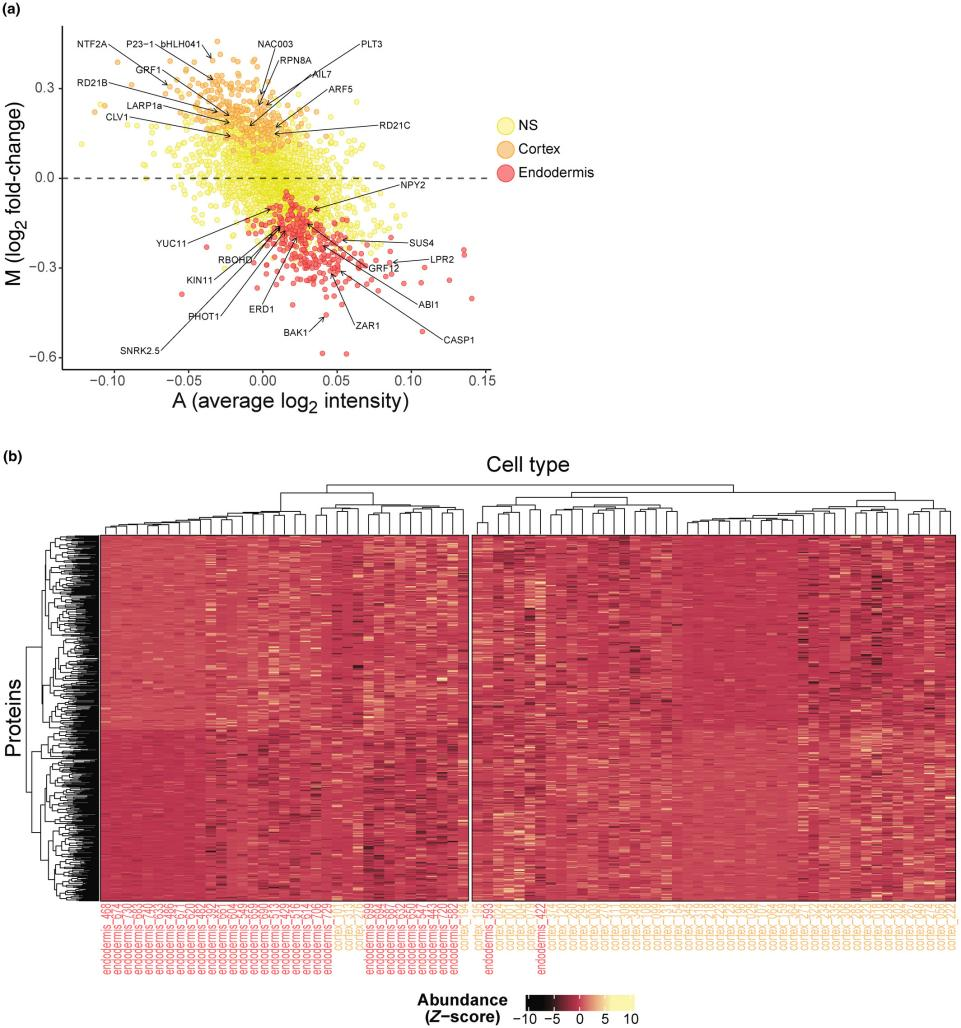
Figure 4. Differential Expression Analysis
In cortical cells, proteins such as AUXIN RESPONSE FACTOR 5, which drives cortical cell division, were enriched. Co-chaperone protein P23-1, critical for auxin distribution and cortical cell maintenance, was also abundant, along with PROTEASOME 19S REGULATORY PARTICLE 8A, which enhances boron tolerance by stabilizing BRAHMA. Hierarchical clustering of cell types and proteins revealed distinct proteomic profiles for endodermal and cortical cells (Fig. 4b).
Development in sample preparation, liquid chromatography, and mass spectrometry have significantly expanded the detection capabilities of single-cell proteomics, enabling the identification of thousands of proteins compared to only hundreds in the past. Building on these developments, the research team at Iowa State University successfully applied single-cell proteomics to analyze cell-type-specific protein expression profiles in Arabidopsis roots. Their findings demonstrated that single-cell proteomics can distinguish different cell types in Arabidopsis roots, confirming the feasibility of this approach in plants and uncovering the intricate protein regulatory mechanisms underlying cellular diversity in plant roots.
MtoZ Biolabs, a leading provider specializing in bioproduct characterization and multi-omics mass spectrometry analysis, is dedicated to delivering expert single-cell proteomics analysis services. Leveraging cutting-edge technology and extensive expertise, we offer robust data support for plant research, including detailed analysis of plant cell types and precise quantification of protein expression variations in complex biological systems. Our tailored solutions are designed to meet the specific requirements of your research, empowering significant advancements in scientific discovery.
References
[1] Montes C, Zhang J, Nolan TM, Walley JW. Single-cell proteomics differentiates Arabidopsis root cell types. New Phytol. 2024 Dec;244(5):1750-1759. doi: 10.1111/nph.19923. Epub 2024 Jun 24.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







