Edman Degradation Sequencing Service
Edman Degradation Sequencing is a classical method of protein sequencing that enables the determination of amino acid sequences from the N-terminus of a polypeptide chain. This technique remains a cornerstone in protein chemistry and structural biology for its precision and reliability in identifying amino acid residues sequentially. Unlike mass spectrometry-based methods, Edman degradation provides direct sequencing information, which is invaluable for validating protein structures, identifying post-translational modifications, and characterizing novel proteins. MtoZ Biolabs offers advanced Edman Degradation Sequencing Services, leveraging this classical yet highly reliable method to provide accurate and detailed protein sequence information essential for your research and development endeavors.
Technical Principles
Edman Degradation operates through a cyclic process that sequentially removes one residue at a time from the amino end of a peptide. The method involves the following key steps:
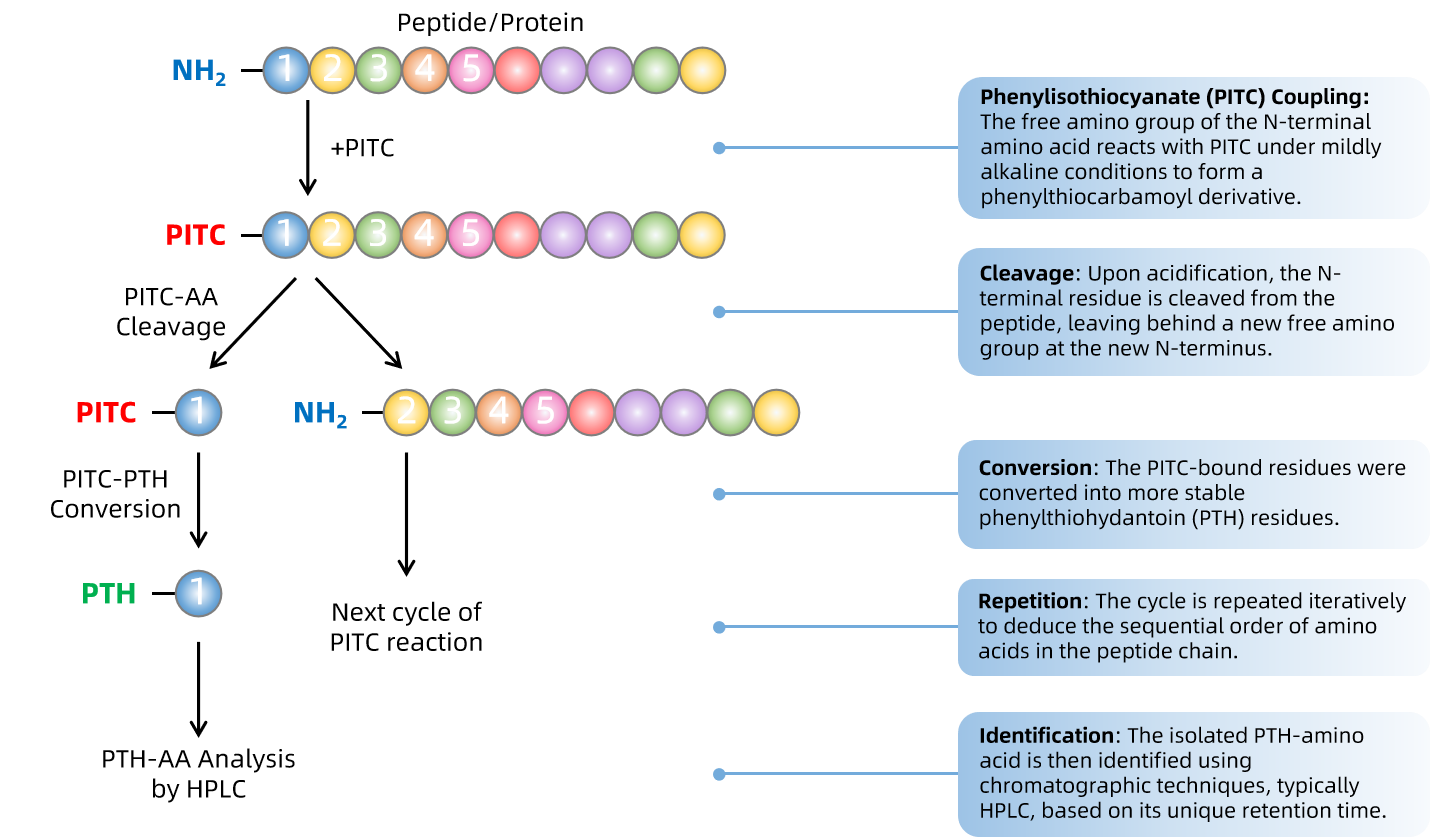
The precision of Edman Degradation relies on the efficient coupling of PITC, the selective cleavage of the PTH-amino acid, and the accurate identification of each amino acid derivative. Typically, the method is most effective for peptides up to 30 amino acids in length, although advancements have extended its applicability to longer sequences under optimized conditions.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. MtoZ Biolabs has developed a robust Edman Degradation sequencing platform leveraging advanced technologies. We offer comprehensive Edman Degradation Sequencing Services for a wide range of purified protein products, including antibodies, protein vaccines, recombinant collagens, and more, tailored to the needs of researchers. Our advanced sequencing system is capable of determining the sequence of up to 30 N-terminal amino acids with complete accuracy. Additionally, with our specialized protein sampling methodology, we can extend this capability to analyze sequences of 60-70 N-terminal amino acids. Welcome to contact to learn more about our service!
Service Advantages
1. Capable of N-terminal sequencing for most proteins with complementary mass spectrometry.
2. Offers high sensitivity for detecting PTH-AA at low picomole levels.
3. Analyzes up to 30 N-terminal amino acids, expandable to 60-70 with optimization.
4. Utilizes the Latest HPLC system equipped with UV-Vis SPD-20A detector for superior separation and detection.
5. Accurately distinguishes similar amino acids, such as I/L and Q/K.
6. Our pricing is transparent, no hidden fees or additional costs.
Applications
1. Sequencing of N-Terminus for Proteins, Peptides, Antibodies, and Vaccines
2. Confirmation of Accurate Translation in Recombinant Proteins
3. Validation of Synthesized Peptide Sequences
Sample Submission Suggestions
1. Electroblotted Samples
Proteins separated by SDS-PAGE should be blotted onto PVDF membrane. Nitrocellulose is not recommended. PVDF membrane can be stained with Coomassie blue or Poncaue red (Sliver stain is not recommended), followed by washing with ultra-pure water. The washing steps must be repeated several times, when glycine-buffer is used.
2. Liquid Samples
1-10 ug sample amount, with >90% purity. Avoid using Tris, glycine, guanidine, glycerol, sucrose, ethanolamine, SDS, Triton, X-100, Tween, and ammonium sulfate in the buffer.
*Note: All reagents/solvent used must be of the highest purity to reduce contaminating substances. Customers are welcome to contact us for detailed sample requirements before sending your samples.
Deliverables
1. Experimental Procedures
2. Parameters of Edman Degradation Sequencing System
3. PTH-AA HPLC Raw Data Files
4. Detailed information on Edman degradation N-terminal sequence analysis
How to order?







