Introduction to Single-Cell Proteomics
Single-cell proteomics refers to the comprehensive and precise analysis of the proteome within individual cells through mass spectrometry, aiming to reveal protein expression patterns under various physiological and pathological conditions. In living organisms, cellular functions are dynamic and are regulated by multiple factors. Even within the same tissue, protein expression patterns can differ significantly among cells. This cellular heterogeneity is a fundamental driver of many biological phenomena. The greatest advantage of single-cell proteomics lies in its ability to analyze cell-type-specific differential protein expression, uncover cellular heterogeneity, and study cellular behavior and function at a finer resolution. This approach enables a clearer understanding of dynamic cellular changes under specific physiological, pathological, or therapeutic conditions. By precisely analyzing the protein composition of individual cells, researchers can deeply investigate intracellular signaling pathways, protein-protein interactions, and metabolic networks, as well as uncover disease mechanisms at the single-cell level. For example, protein heterogeneity inside and outside tumor cells can influence tumor invasiveness, metastatic potential, and therapeutic response. Therefore, single-cell proteomics has become an essential tool for studying personalized cancer treatments. As a professional provider of mass spectrometry services, MtoZ Biolabs is dedicated to offering one-stop single-cell proteomics solutions, helping researchers efficiently reveal complex protein networks within individual cells.
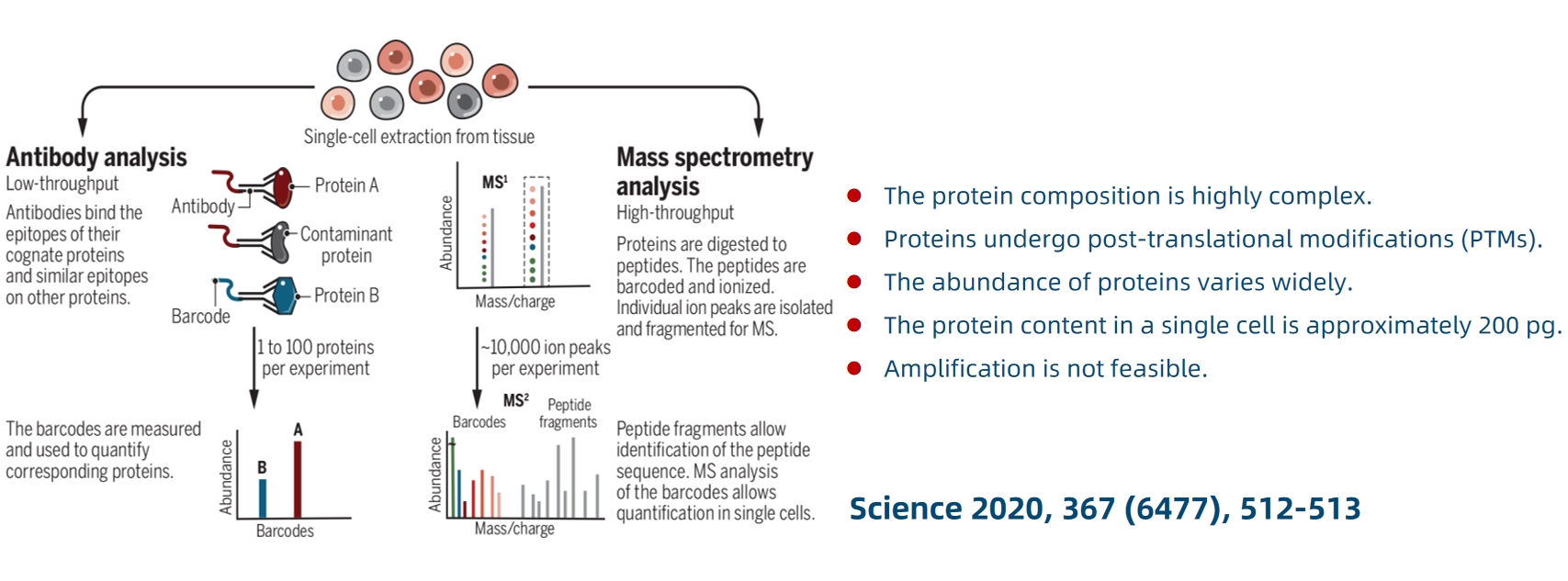
Figure 1.
Why Conduct Single-Cell Research?
1. Single-cell research provides insights into the roles of distinct cell subpopulations within the tumor microenvironment, advancing our understanding of cancer initiation, progression, metastasis, and immune evasion, and offering potential therapeutic targets.
Tumors comprise diverse cell types, including cancer cells, fibroblasts, immune cells, and endothelial cells, each with distinct subpopulations. The intricate interactions between these cells often drive cellular transformations within the tumor microenvironment, promoting tumor growth. For example, fibroblasts are reprogrammed to provide physical and biochemical support to cancer cells, while vascular development ensures an adequate supply of nutrients and oxygen. Immune cells, which typically suppress tumor growth, are frequently co-opted by cancer cells to promote their proliferation. Given the high heterogeneity of these cell types, studying the roles of specific subpopulations in cancer processes or identifying molecular targets for innovative therapies requires single-cell analysis.
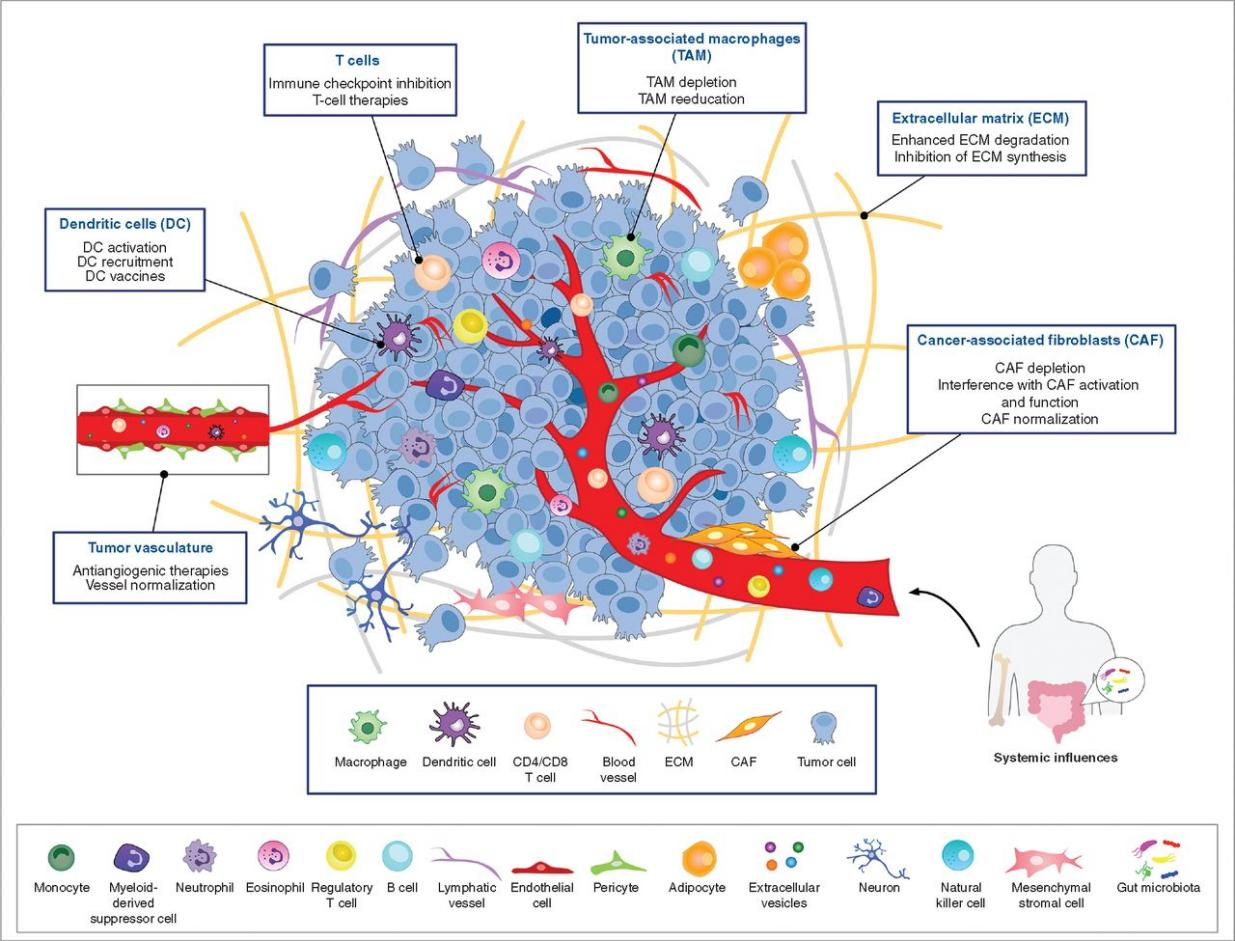
Figure 2. Cancer Discov (2021) 11 (4): 933-959.
2. Single-cell analysis represents a pivotal advancement in precision medicine.
3. Single-cell omics technologies demonstrate substantial advantages over traditional omics approaches.
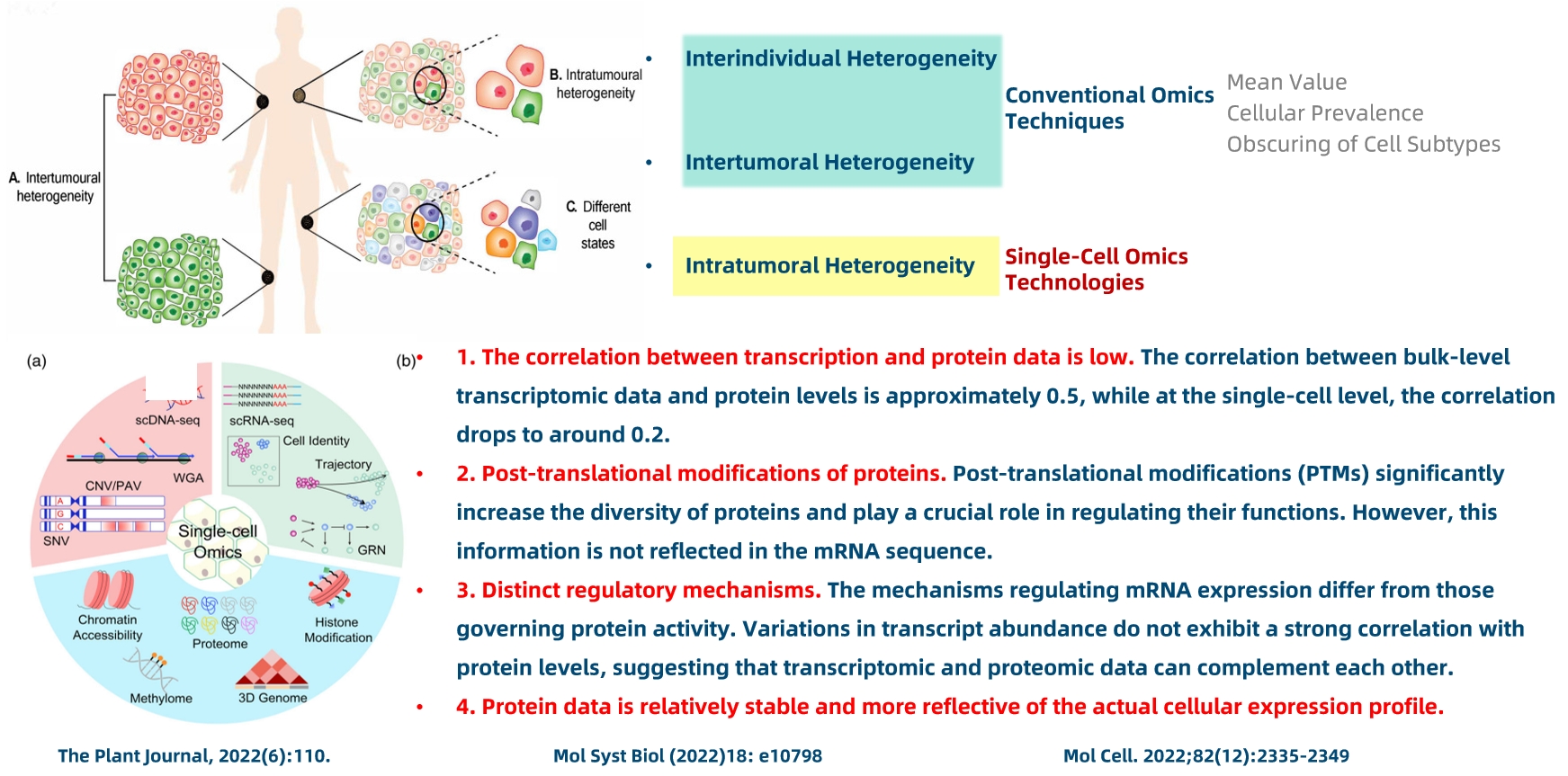
Figure 3.
4. Single-cell proteomics provides a more detailed and accurate characterization of cellular properties.
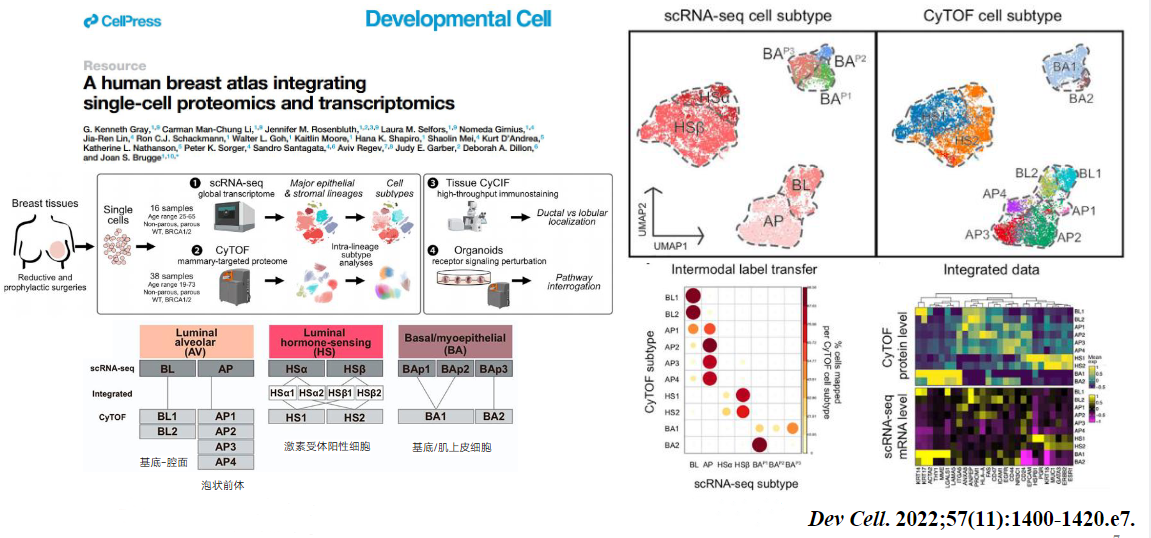
Figure 4.
Analysis Workflow of Single-Cell Proteomics - CellenOne
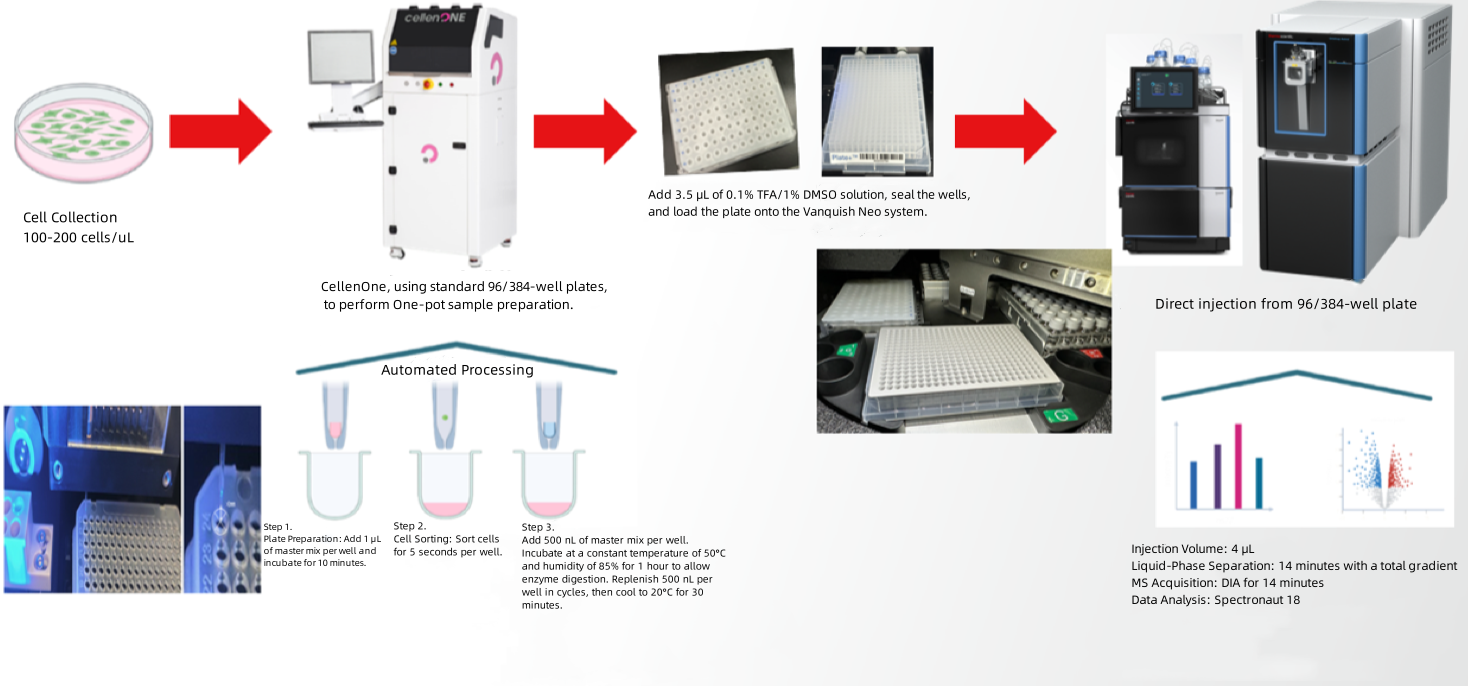
Figure 5.
1. Principles of CellenOne Technology
CellenOne employs an automated mechanical arm to position a sample needle containing a cell suspension in front of a high-speed camera. The needle's piezoelectric device generates controlled droplets, which are analyzed in real time. When a droplet contains a single cell meeting predefined parameters-such as diameter, roundness, and fluorescence intensity-it is directed into a well plate. Droplets containing multiple cells or no cells are returned to a recovery tube for redistribution.
2. Advantages of CellenOne Technology
(1) Gentle Sorting: Piezo-acoustic dispensing minimizes shear stress and cell loss, enhancing viability and compatibility across diverse cell types.
(2) Optical Monitoring: Advanced visual processing integrates software-based feedback to ensure precise sorting.
(3) High Single-Cell Accuracy: Guarantees accurate single-cell isolation, achieving a success rate of up to 98%.
(4) Reduced Loss Rates: Droplets with multiple cells are recycled for redistribution, minimizing waste.
(5) Full Automation: A fully automated workflow combines mechanical arms with nano-scale reagent dispensing systems, ensuring reproducibility and streamlining the process from suspension to single-cell preparation.
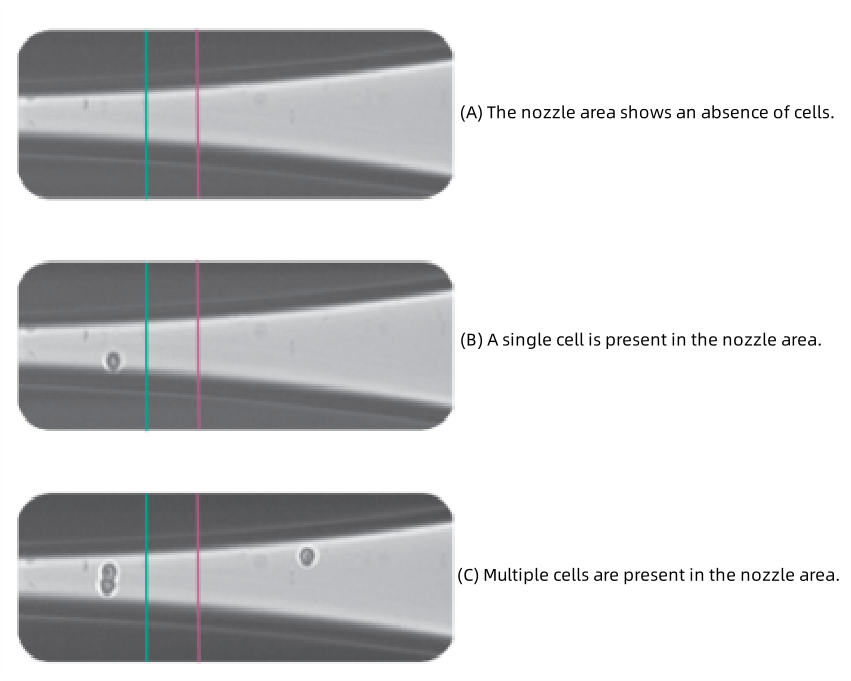
Figure 6.
Examples of Single-Cell Proteomics Analysis
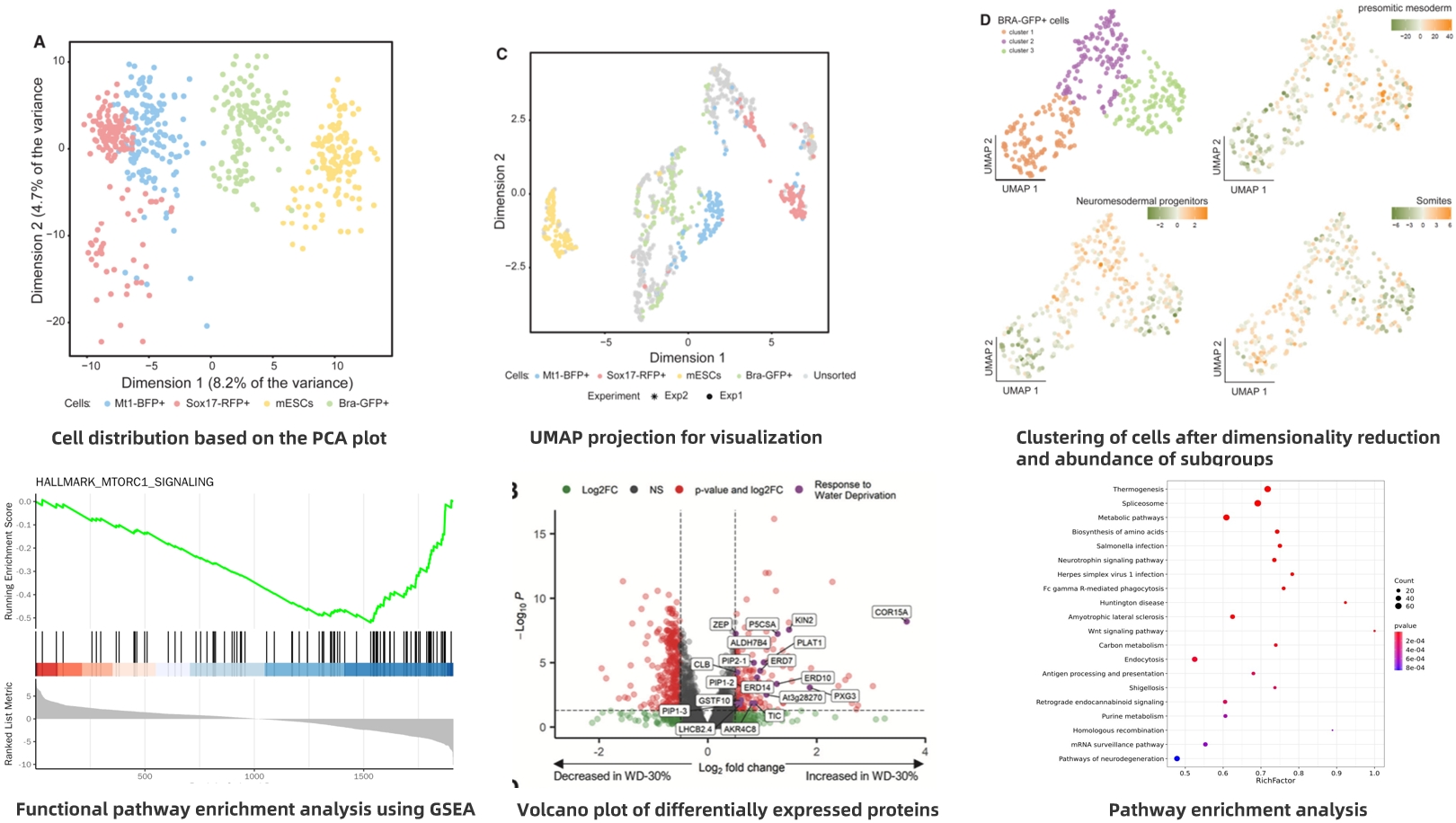
Figure 7.
References and Applications of Single-Cell Proteomics
Single-cell proteomics is particularly effective in contexts with limited cellular heterogeneity, such as early embryonic development and stem cell differentiation.
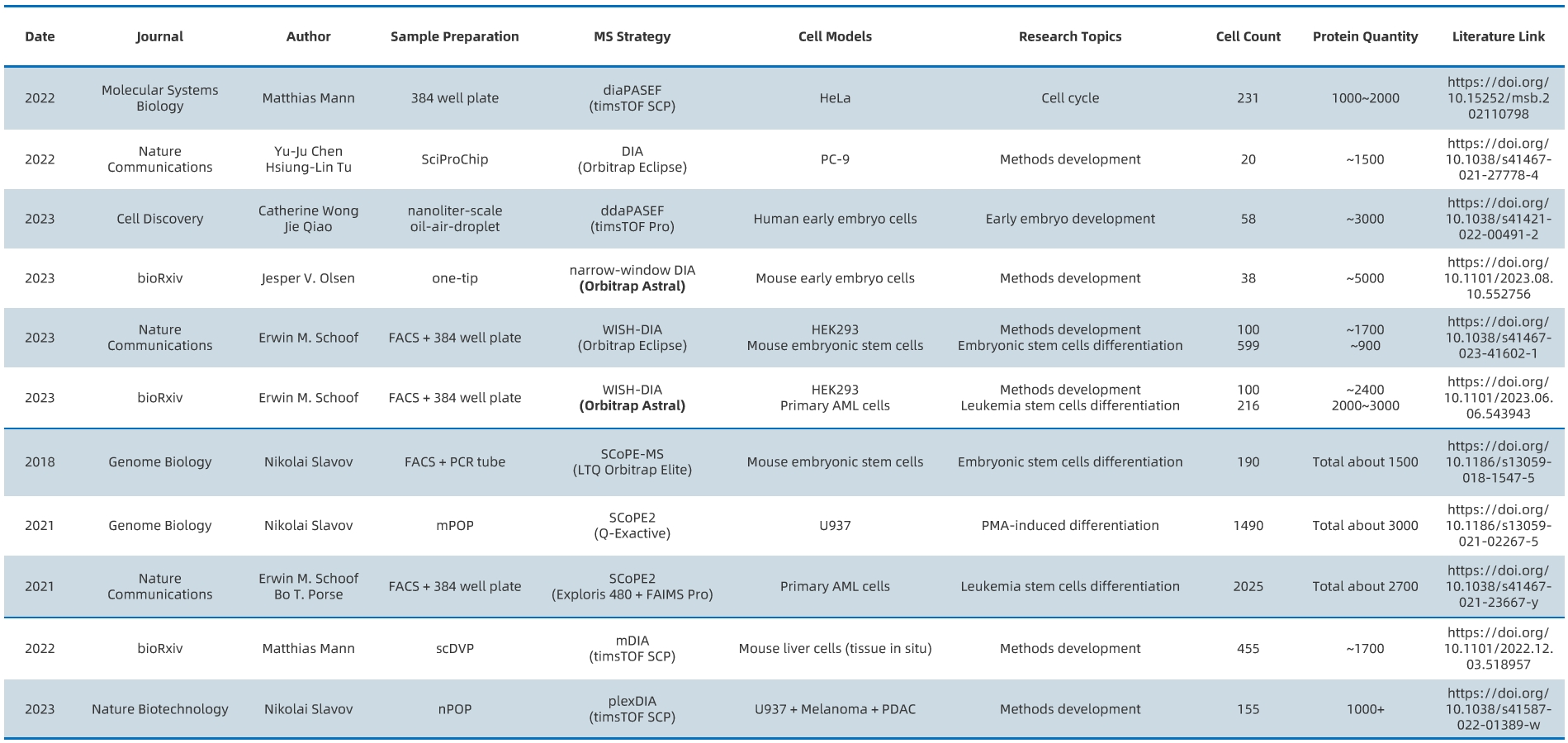
Figure 8.
Application Case 1: Deciphering Cellular Heterogeneity in Early Embryogenesis
Title: Deciphering Lineage Specification During Early Embryogenesis in Mouse Gastruloids Using Multilayered Proteomics
(1) Scientific Question: The process of germ layer formation during embryonic development involves complex gene regulatory programs, yet proteomic studies on early mammalian development remain relatively limited.
(2) Sample: Mouse embryo-like structures derived from embryonic stem cells (mESCs), including SOX17-RFP+ (endoderm), BRA-GFP+ (mesoderm), MT1-BFP+ (ectoderm) cells, and undifferentiated mESCs.
(3) Omics Approaches: Single-cell proteomics, bulk proteomics, single-cell transcriptomics, CHIP-seq
(4) Key Findings:
① Quantified 560 individual cells and identified 2259 proteins.
② Single-cell proteomics analysis of BRA-GFP+ cells (363 cells) revealed three subpopulations corresponding to neural mesoderm precursors (NMPs), somitic mesoderm, and presomitic mesoderm (PSM) subtypes.
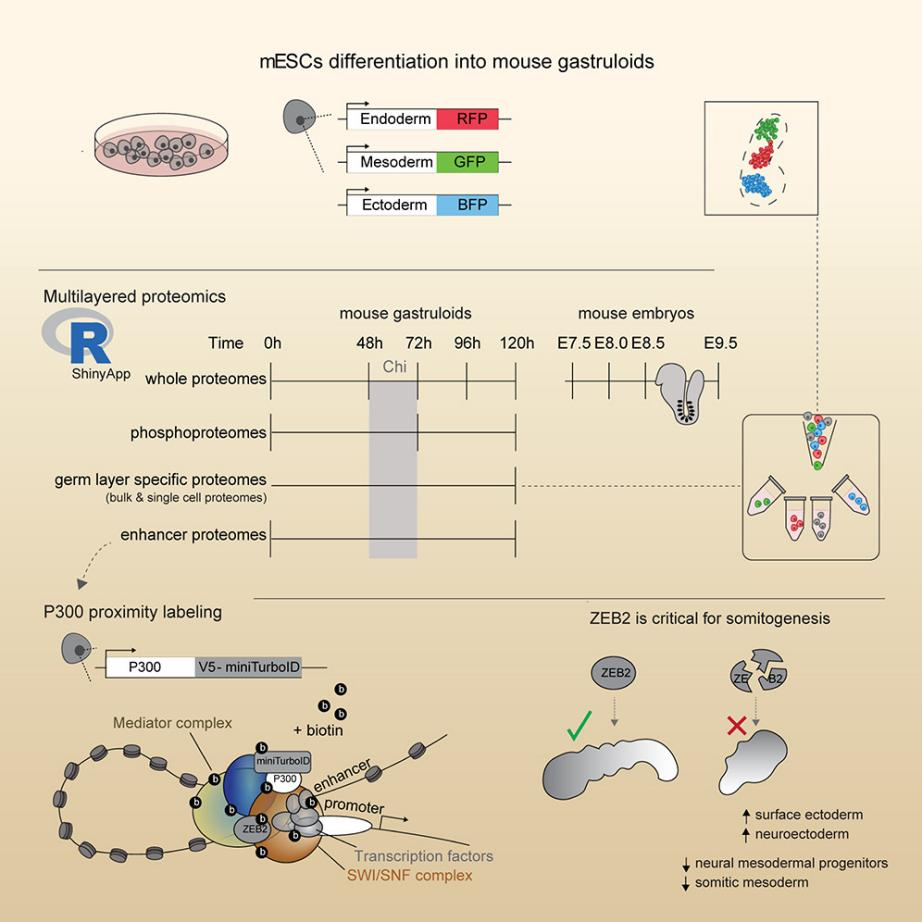
Figure 9. Cell Stem Cell. S1934-5909(24)00144-9. 10 May. 2024
Application Case 2: Tracing Drug Resistance Trajectories
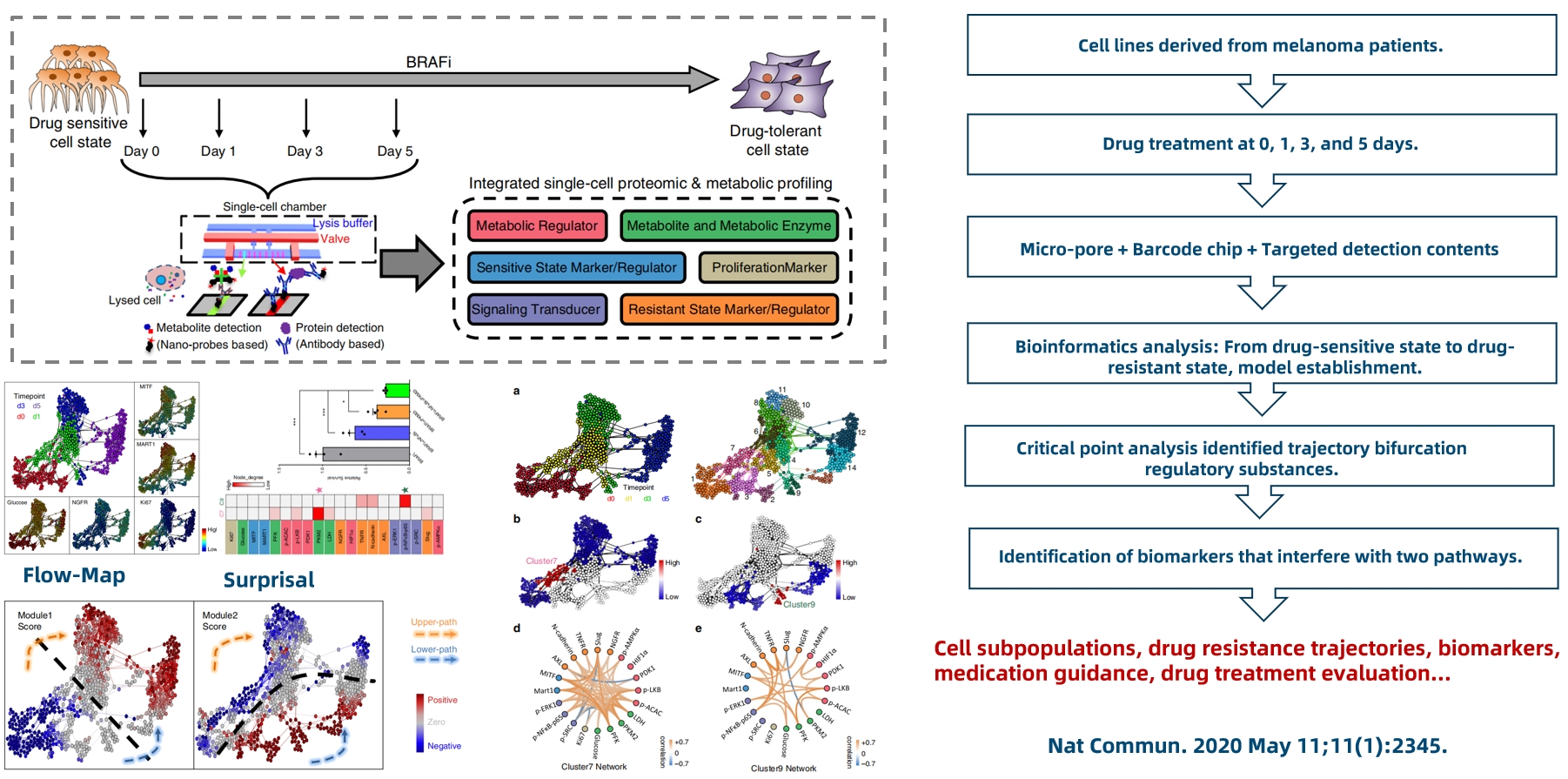
Figure 10.
Application Case 3: Exploring Mechanisms of Drug Response
Title: Single-Cell Chemical Proteomics (SCCP) Interrogates the Timing and Heterogeneity of Cancer Cell Commitment to Death
(1) Scientific Question: How do individual A549 adenocarcinoma cells respond to three anticancer drugs (Methotrexate MTX, Camptothecin CPT, and Tomotidine TDX) at the single-cell level?
(2) Sample: A549 cells treated with drugs at specific time points, with 96 cells analyzed per time point.
(3) Omics Approach: Single-cell proteomics
(4) Key Findings:
① Protein expression changes were tracked in MTX-treated cells, distinguishing two subpopulations (G1 and G2) predictive of future cell death or survival, reflecting response heterogeneity.
② Comparative analysis with controls identified drug targets and monitored cellular responses at the single-cell level.
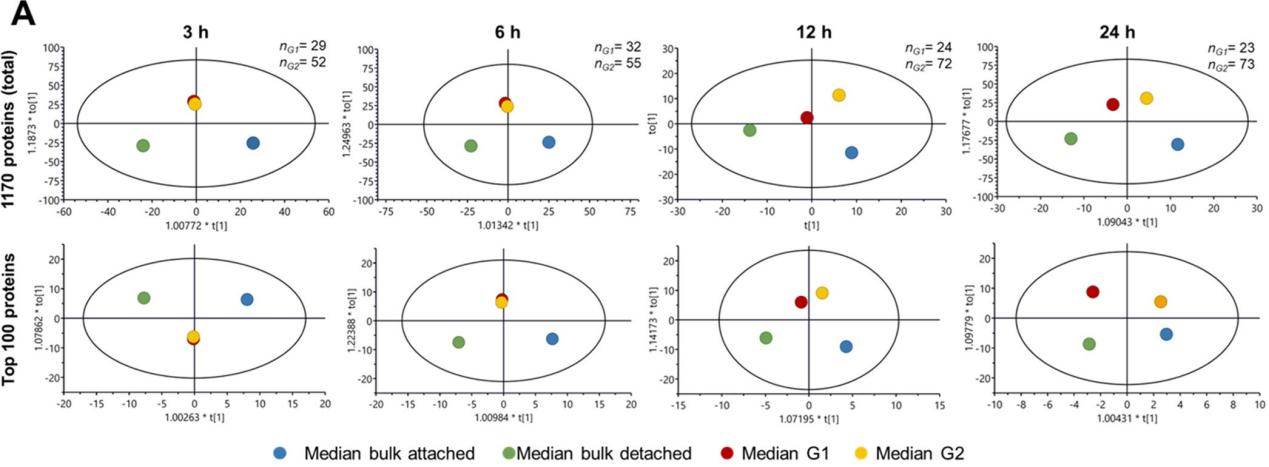
Figure 11.
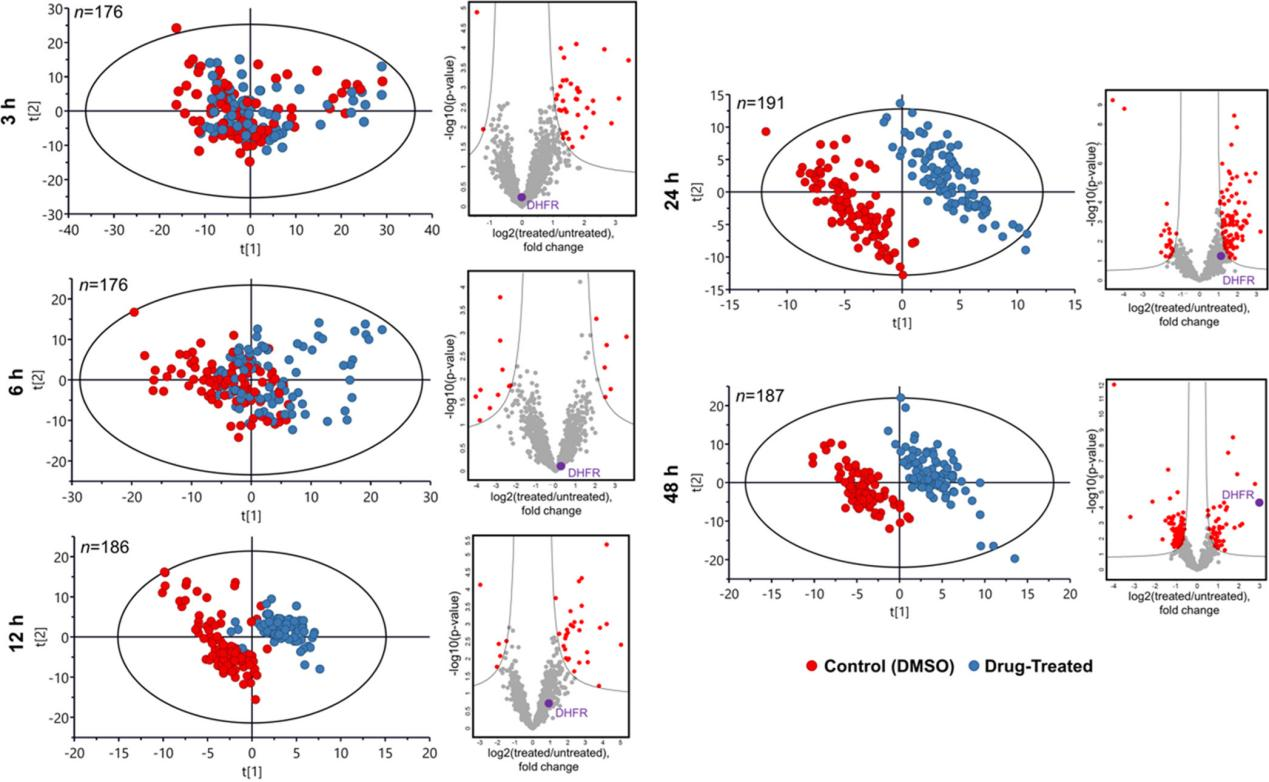
Figure 12.
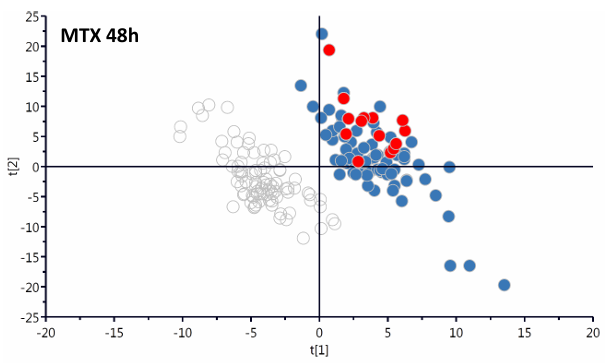
Figure 13. Anal Chem. 2022;94(26):9261-9269.
Application Case 4: Differentiating Plant Cell Types
Title: Single-Cell Proteomics Differentiates Arabidopsis Root Cell Types
(1) Background: Single-cell proteomics (SCP) offers a novel approach to unraveling cellular heterogeneity in multicellular organisms. This study demonstrates SCP’s applicability to plant samples using Arabidopsis as a model.
(2) Sample: Isolated single cells from the cortex and endodermis, two neighboring root cell types derived from a shared stem cell lineage.
(3) Omics Approach: Single-cell proteomics
(4) Key Findings:
① From 756 root cells, 3763 proteins were identified, with an average of 1118 proteins per cell. After stringent filtering, 3217 proteins were reliably quantified.
② A subset of 596 proteins exhibited enrichment in either the cortex or endodermis, effectively distinguishing these closely related cell types.
The findings highlight SCP’s capability to resolve adjacent cell types with functional differences, enabling the discovery of biomarkers and candidate proteins.
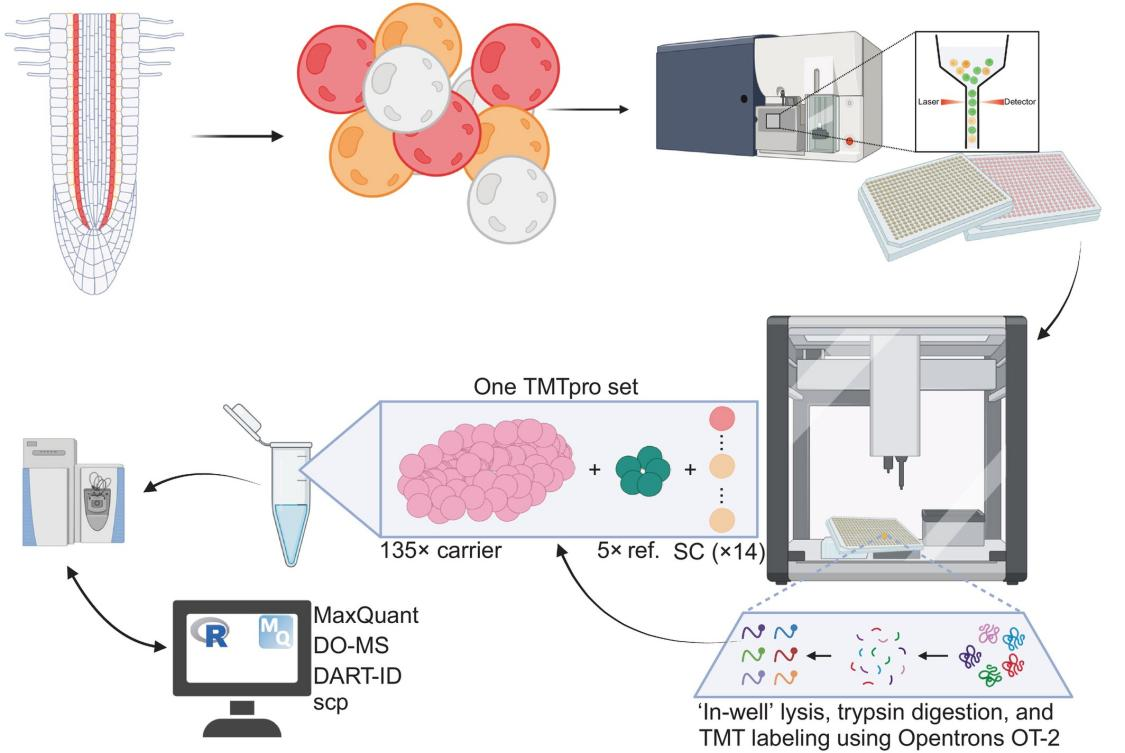
Figure 14.
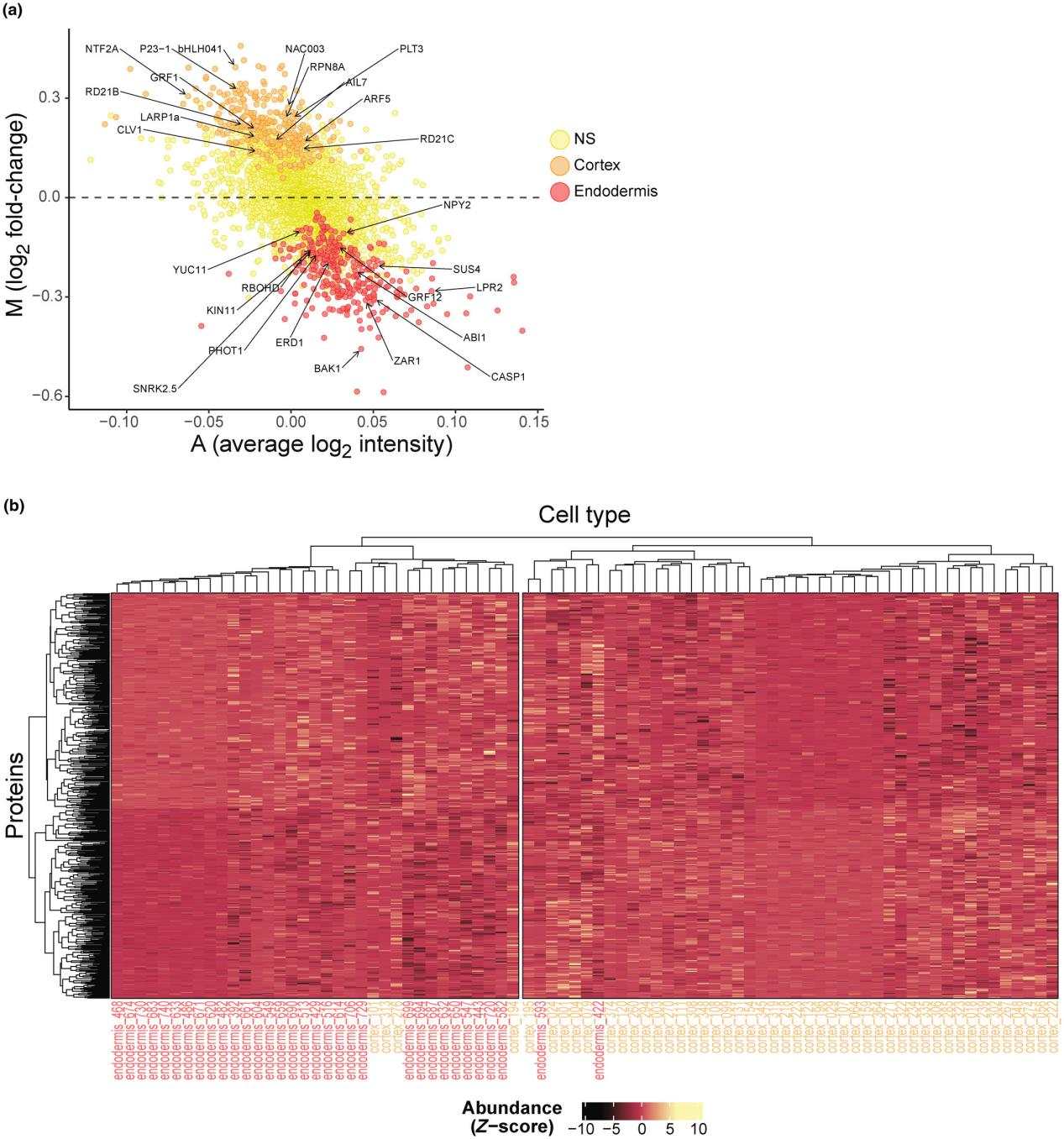
Figure 15. New Phytol. 2024 Dec;244(5):1750-1759.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







