LC-MS/MS of Protein C-terminal Sequence Determination Service
The C-terminal sequence is a pivotal structural and functional part of proteins and peptides, which may contain significant antigenic determinants (i.e., parts that trigger an immune response). Analysis of these regions' sequences can help researchers identify which fragments might be most useful in vaccine design. Additionally, analysis of the C-terminal sequence may reveal that certain regions play a crucial role in the pathogen's lifecycle or infection process, such as bacterial adhesion or viral fusion and entry into host cells. Due to the need for further cleavage and modification of proteins post-translation within the cell, it is often not feasible to simply use the gene sequence to predict the C-terminus straightforwardly. More accurate methods, such as mass spectrometry, are required to confirm the position of the C-terminus. In the analysis of recombinant protein expression and purification products, especially in the development and process establishment of recombinant protein products, it is also necessary to confirm the C-terminal sequence of proteins to ensure the stability of the entire process.
MtoZ Biolabs has established a protein C-terminal sequence analysis service tailored to different types of bioproducts based on the principles of peptide mapping analysis (mass spectrometry). This service enables the determination of the C-terminal sequences of bioproducts such as proteins, antibodies, vaccines, peptides, and recombinant collagen.
Currently, a technique for determining the C-terminal sequence of proteins similar to Edman degradation has not been developed, thus services for verifying the C-terminal sequence of proteins are still based on mass spectrometry platforms. According to the theoretical sequence information of the target protein, two complementary proteases were selected to produce two C-terminal peptides with different lengths but suitable for ionization. After mass spectrometry detection and mutual verification, the C-terminal sequence of the protein was finally determined.
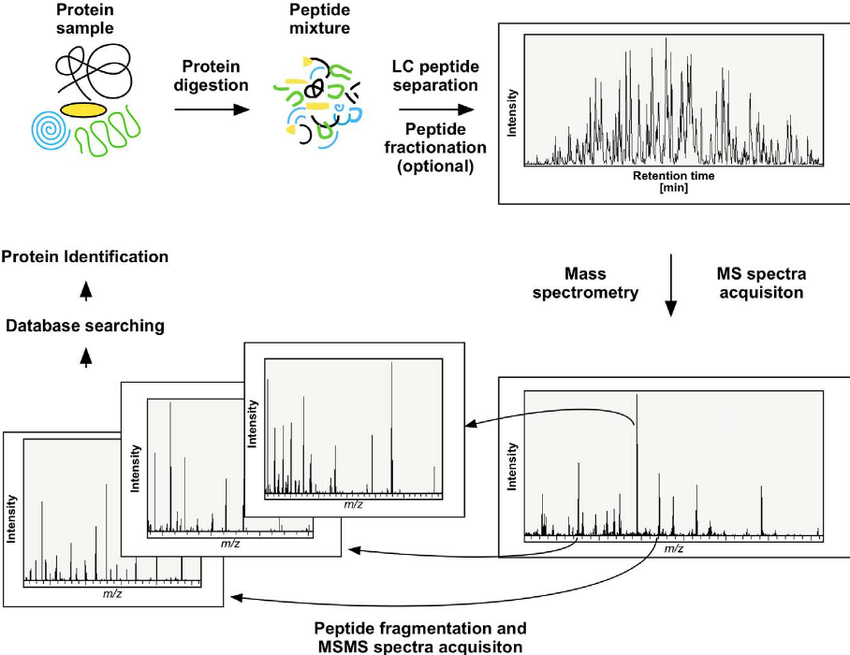
Analysis Workflow
1. Protein samples are enzymatically digested into peptides.
2. Desalted peptides are analyzed with a Q-Exactive mass spectrometer (Thermo Fisher Scientific).
3. Spectra matching against theoretical sequences identifies the protein's C-terminal sequence.
Service Advantages
1. Employ Specialized Software for Matching Mass Spectrometry Data with Theoretical Sequences
2. Determine the Specific Location of the C-Terminus in the Protein Sample
3. Provide a Secondary Mass Spectrum of the C-Terminus
Sample Results
After appropriate enzymatic digestion into peptides, the protein sample undergoes mass spectrometry analysis, resulting in the following spectral output.
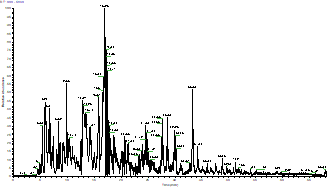
The Total Ion Chromatogram (TIC) of The Sample
The horizontal axis represents time (min) and the vertical axis represents relative signal intensity.
The molecular weight of the complete peptide can be obtained by the MS1 spectrum, mass = (m/z-1) * z.
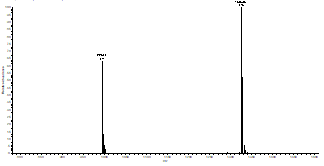
Peptide Segment MS1 Spectrum
The horizontal axis represents detected m/z values, and the vertical axis represents relative signal intensity.
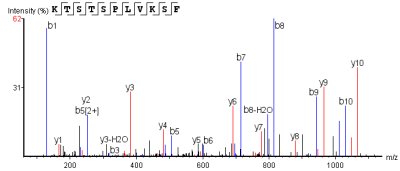
Peptide Segment MS2 Spectrum
The horizontal axis represents detected m/z values, and the vertical axis represents relative signal intensity.
Sample Submission Requirements
1. Suitability for Gel and Solution Samples
2. Target Protein Sample Requirement: 5-10ug
3. Preference for Higher Sample Purity
Services at MtoZ Biolabs
1. Project Report, Including
(1) Experimental procedures.
(2) Relevant mass spectrometry parameters.
(3) Detailed information on protein C-terminal sequences.
(4) MS Spectrum, etc.
2. Raw Data
FAQ
Q1: Protein after digestion will be broken at a fixed site, how to determine the c-terminal peptide is the protein c-terminal?
During data analysis, peptide segments where the N-terminus is the cleavage site while the C-terminus is not are selected to ensure that the peptide segments are formed by cleavage rather than random impurities. At the same time, it is ensured that the end of the peptide segment matches the reference end.
Q2: How to confirm the specific position of the C-terminus if the analysis results only have peptides with the C-terminus as the enzyme cleavage site?
Such outcomes typically arise from two distinct situations.The first case is that the C-terminal of the protein happens to be the cleavage site, and we can eliminate this case by different digestion at the same time. The second case occurs when the C-terminus of the protein is either very close to or far from the enzyme cleavage site, resulting in fragments that are either too short or too long after enzymatic cleavage. The fragmentation is too poor to be recognized by analysis software. To address this, we can establish a gradient database in which each sequence has one more amino acid than the previous sequence. This gradient database can then be used for re-analysis through alignment.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
N/C Terminal Sequencing Service
How to order?







