Micro-Crystal Electron Diffraction Analysis Service
Microcrystal electron diffraction (MicroED) is a novel technique that uses cryo-transmission electron microscopy (Cryo-TEM) for the three-dimensional (3D) reconstruction of organic small molecules and biological macromolecule crystals based on electron diffraction data. As a new technology, MicroED employs electrons as the incident light beam to acquire diffraction data from micrometer and nanometer-scale 3D crystals at ultra-low electron dose rates. During data acquisition, crystals continuously rotate on the sample stage, and a high-speed camera records the diffraction patterns. Like X-ray crystallography, the samples must be crystallized, although the crystal size required is significantly smaller, only at the nanometer scale. MicroED data can be processed using standard X-ray crystallographic software to generate 3D models. The main advantage of MicroED is the ability to diffract and extract high-resolution structural information from samples that resist forming large crystals while using significantly less material. Unlike X-rays, which only produce electron density maps, electrons can probe the electrostatic potential of substances, allowing MicroED to map the charged states of atoms within the structure.
MicroED combines the strengths of cryo-EM and X-ray crystallography, facilitating rapid, high-resolution determination of crystal structures of organic molecules and biomolecules. The advantages of the two technologies complement each other and empower the structure biology research. To date, the resolution of MicroED structure determinations has been relatively high, fully demonstrating the powerful capabilities of this technology.
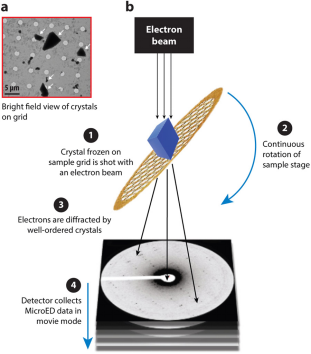
Figure 1. Overview of MicroED [3]
The MicroED workflow begins by preparing the crystal and placing it on an electron microscope grid. The grid is then inserted into an electron microscope where the electron beam is aligned with the microcrystal. Screening for electron diffraction quality is conducted manually or automatically. Once well-diffracting crystals are identified, they are continuously rotated to collect comprehensive electron diffraction data. This data is processed using standard X-ray crystallography software to refine and generate precise crystal structures.
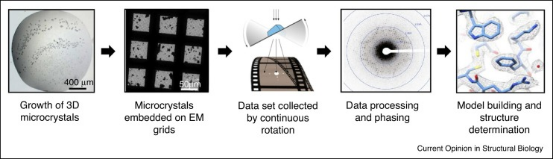
Figure 2. Experimental Flow of MicroED [4]
1. Crystal Preparation and Screening
The preparation of protein crystals for MicroED follows the same protocols as X-ray crystallography but requires significantly fewer crystals. However, MicroED can extract structural information from crystals that are one-billionth the size required for X-ray diffraction. Identifying MicroED microcrystals is challenging under light microscopes, necessitating alternative detection techniques such as ultraviolet (UV) fluorescence, second-order nonlinear imaging of chiral crystals (SONICC), fluorescence microscopy, and negative-stain electron microscopy.
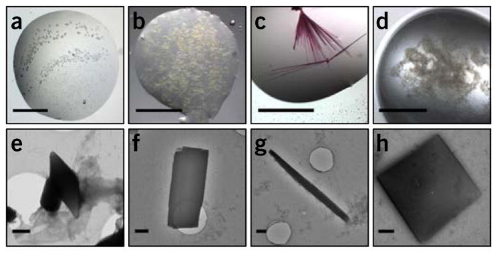
Figure 3. Representative Images of Protein Microcrystals [5]
2. MicroED Sample Preparation
After retrieving the microcrystals, they are placed on a glow-discharged electromagnetic grid and dried to remove excess solution before being cryo-cooled in liquid ethane. For anhydrous samples, such as small molecules or natural products, the crystals, often as powder, are directly placed on the grid and analyzed under cryogenic conditions to minimize radiation damage. In some cases where the samples are robust, MicroED analysis can be conducted without cooling. If larger crystals (1–5 μm) form, they are still challenging for traditional X-ray crystallography but are fragmented or processed using FIB-SEM to produce smaller crystals suitable for electron diffraction.
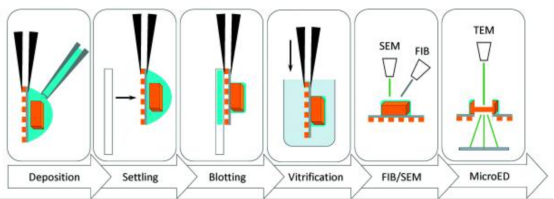
Figure 4. Typical Workflow for MicroED Sample Preparation of Protein Crystals [6]
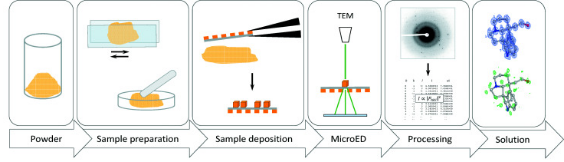
Figure 5. Typical Workflow for MicroED Sample Preparation of Small Molecule Powders [6]
3. Data Screening and Collection
After the crystals are placed on the grid, it is loaded into the TEM under cryogenic conditions to begin data collection. The grids are generally evaluated in bright-field mode or overfocused diffraction to assess the overall thickness of the ice layer and to identify areas containing crystals of appropriate size. Once the crystals are located, test diffraction patterns are recorded to assess their quality. This step can be automated. Once a region with crystals is identified, the ice thickness is minimized based on the contrast between the carbon support and the holes, and the microscope switches to overfocused diffraction mode. In this setting, individual crystals are detected, and the z-height is finely adjusted while keeping the dose rate below 10^-3 e Å^-2 s^-1, and the central spot is precisely focused by minimizing the size of the direct beam spot. High-quality diffraction patterns are recorded on a fast camera as sequences, with the electron dose rate (exposure) set to 0.01–0.05 e^-Å^-2s^-1 while the crystal continuously rotates on the sample stage. If high-quality diffraction is observed, a complete dataset is collected from that crystal. The cryo-holder's edges limit the tilt range to approximately ±70°, with the grid perpendicular to the electron beam.
In shutterless mode, continuous rotating datasets from a single crystal are collected within about 10 min. The detector is continuously exposed, and data are read out at regular intervals. This mode of operation, which sacrifices detector precision for simplicity in the experimental setup, has been found to be effective. Notably, the impacts of intensity accumulation and uninterrupted sample rotation during the detector's ∼0.1 s readout time are negligible. The exposure used in MicroED is about 100 times lower than other cryo-EM modes, significantly reducing radiation damage. Continuous rotation of the sample in MicroED enhances the accuracy of reflection intensities and reduces the effects of dynamic scattering, making these datasets superior to those obtained from stationary diffraction.
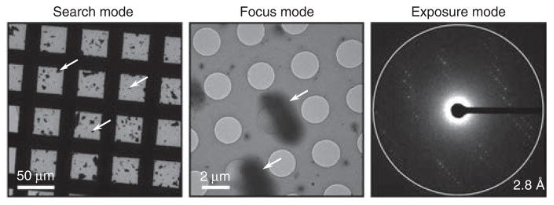
Figure 6. Example Images of Search, Focus, and Exposure Modes from a Model MicroED Sample [5]
4. Data Processing
During MicroED diffraction, crystals rotate continuously at a constant angular velocity, similar to the method used in X-ray crystallography. Each frame of the MicroED sequence captures a wedge of reciprocal space. Therefore, MicroED data can be processed directly using conventional X-ray crystallography software. Successful indexing, integration, and scaling of MicroED data have been achieved using software such as XDS, iMOSFLM, DIALS, SHELX, and HKL2000. High-resolution data sets (better than ~1.2 Å) are necessary for solving structures from scratch. Historically, most protein structures resolved through MicroED have utilized molecular replacement to address the phase problem, relying on the availability of structurally homologous search models. MicroED requires multiple diffraction patterns with a minimum tilt range of 20° to capture the essential 3D information, enabling successful indexing and determination of crystal orientations without prior knowledge. Following data integration, merging, and scaling, phase analysis and model refinement can be conducted using standard crystallographic software suites such as CCP4 and PHENIX. Apart from high-resolution peptides, small molecules, and metals that can be phased directly, all MicroED structures have been phased through molecular replacement. Refinement typically employs REFMAC or phenix.refine66, following the same protocols used with X-ray data but utilizing electron instead of X-ray scattering factors.
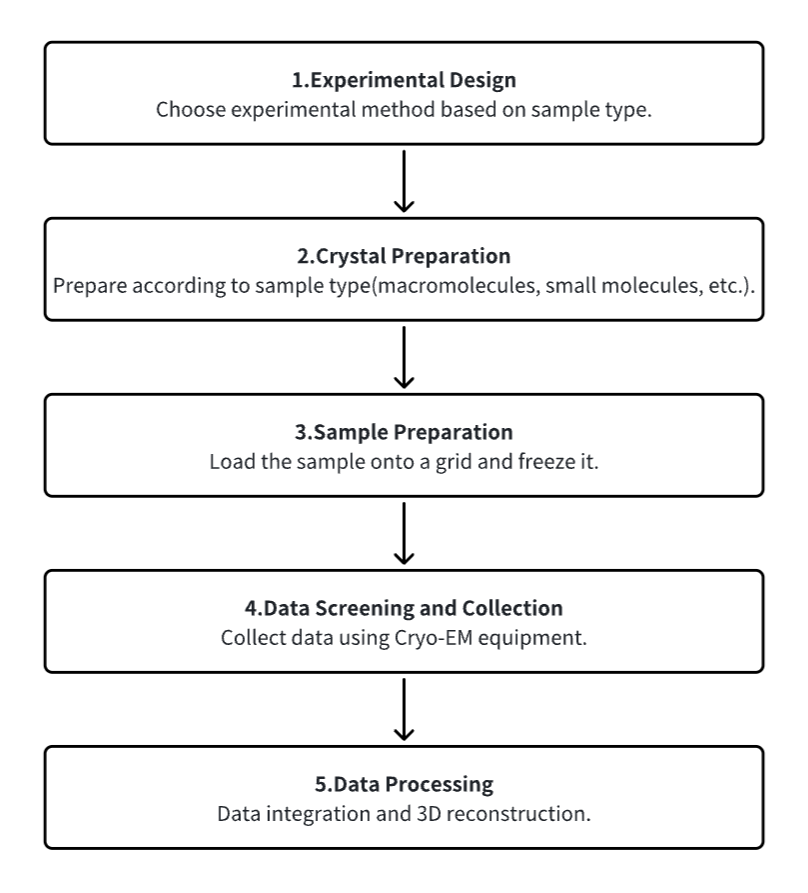
Analysis Workflow
1. Experimental Procedure Determination Based on Requirements
2. Crystal Preparation
3. Sample Pretreatment and Freezing
4. Data Screening and Collection
5. Data Processing
Service Advantages
1. Customizable Experimental Methods Based on Requirements
2. Multi-Gradient Design for Optimal Condition Exploration and Sample Optimization Guidance
3. Automated Sample Preparation Platform for High-Throughput and Rapid Sample Processing; Brief Electron Microscope Session Times; Precise Control of Reaction Conditions in Less than 30 Minutes per Sample
4. High-Resolution Cryo-EM: Image Collection of Crystals Smaller than 200 nm; Molecular Structures at Atomic Resolution Finer than 1 Å
Sample Results
1. Structure Determination of DNA Crystals by MicroED
MicroED serves as a powerful tool for determining the high-resolution structures of microcrystals from diverse biological molecules, chemicals, and materials. This study applied MicroED to previously unanalyzed DNA crystals, using the d(CGCGCG)2 DNA duplex as a model. Cryo-focused ion beam (cryo-FIB) milling was utilized to prepare thin sections for diffraction data collection. The MicroED data collection and subsequent processing resulted in a 1.10 Å resolution structure of the d(CGCGCG)2 DNA, demonstrating the successful application of cryo-FIB milling and MicroED to the investigation of nucleic acid crystals.
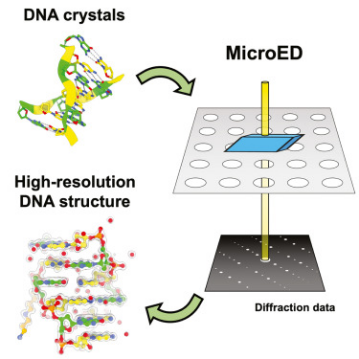
Figure 7. MicroED Determination of DNA Structure [7]
2. Electron Diffraction Determination of the Structure of Levocetirizine Dihydrochloride
Levocetirizine is an orally active second-generation antihistamine drug component, used for treating allergy symptoms and chronic urticaria for over 25 years. Despite its widespread use, its crystal structure remains unknown. A study reported the application of 3D electron diffraction (3D ED)/MicroED to directly determine the crystal structure of Levocetirizine dihydrochloride from crystalline powder extracted from commercially available tablets. The results also demonstrated the practicality of dynamic refinement in explicitly assigning absolute configurations. The results highlight the immense potential of 3D ED/MicroED for structure elucidation of components of microcrystalline mixtures that obviates the need to grow large-size single crystals and the use of complementary analytical techniques, which could be important for identification as well as for primary structural characterization.
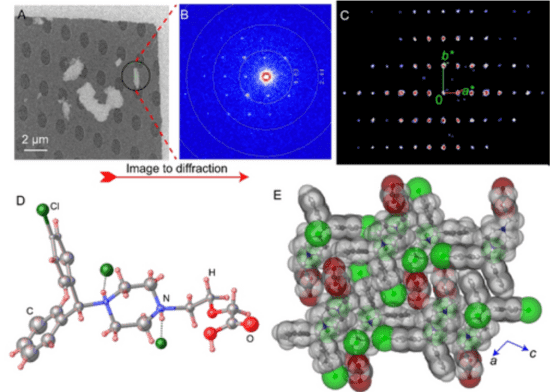
Figure 8. 3D ED/MicroED Measurement and Structural Details of Levocetirizine Dihydrochloride Tablet Crystalline Powder [8]
3. MicroED Structure of the Human Adenosine Receptor Determined from Single Nanocrystals in LCP
MicroED is a cryo-EM technique that enables the determination of protein structures from sub-micrometer crystals. G protein-coupled receptors (GPCRs), crucial drug targets, require crystallization within lipid cubic phases (LCP), which complicates standard MicroED applications. Research demonstrated that GPCR microcrystals grown in LCP could be adapted for MicroED by converting LCP into a sponge phase followed by ion beam milling into thin sections. These results provide a foundation for using MicroED in resolving GPCR structures, with potential applications in structure-based drug discovery.
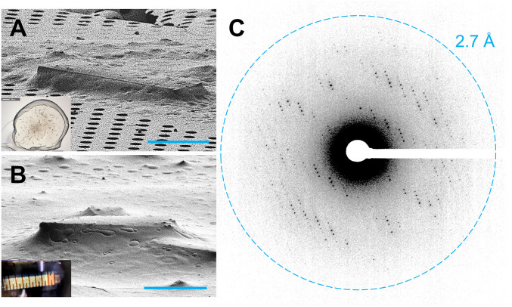
Figure 9. GPCR Crystals Prepared for MicroED Data Collection [9]
4. Characterization of Reactive Organometallic Substances by MicroED
Research has applied MicroED to determine structures of transition-metal complexes. Using 300 keV electrons, very low electron doses, and an ultrasensitive camera enabled data collection without cryo-cooling the sample holder. This technique revealed the first crystal structure of classical zirconocene hydride, commonly known as the "Schwartz's reagent," unsuitable for solution-state NMR or X-ray crystallography, along with five other paramagnetic and diamagnetic transition-metal complexes.
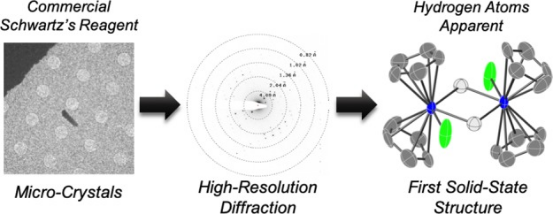
Figure 10. Metal Structures Obtained by Ambient Temperature MicroED Method [10]
Sample Submission Requirements
1. Comprehensive Experimental Steps
2. Specifications of Relevant Instrumentation
3. Original Experimental Data
4. Data Analysis Report
Applications
1. Use of MicroED in Drug Discovery
MicroED offers numerous opportunities to advance challenging drug discovery projects. It has particular strengths in studying small molecule drug candidates, natural products, ligand-protein complexes, and membrane proteins for drug discovery applications. With recent technological advances in this field producing very high-resolution structures, novel protein-ligand complexes, and many new or modified small molecule structures, MicroED has proven to provide critical information and has the potential to become a significant method in future drug discovery.
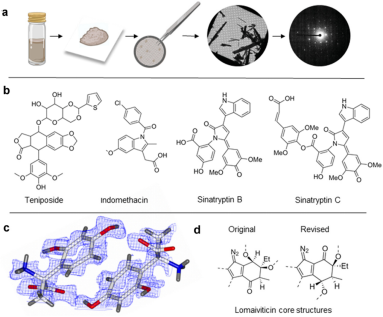
Figure 11. MicroED for Identifying Small Molecule Structures [11]
2. MicroED in Discovering Protein Interactions
Understanding the interactions between small molecules and their protein targets is crucial for foundational studies of ligand binding as well as for drug development and optimization. Although X-ray crystallography is the traditional method for studying the high-resolution structural details of ligand-protein and drug-protein complexes, this method often requires large crystals that can be challenging to produce. This is particularly true for natural products isolated from natural sources, which are usually available in limited quantities insufficient for large-scale crystal growth. Therefore, developing methods that can determine structures using less material and smaller crystals has become particularly important. MicroED technology shows great potential in this area. It can obtain high-resolution structural data from nanocrystals and microcrystals, thereby eliminating the need for large crystal growth. Furthermore, MicroED can improve the results of ligand soaking experiments by using smaller crystals in some cases, thus accelerating the research on protein-ligand complexes.
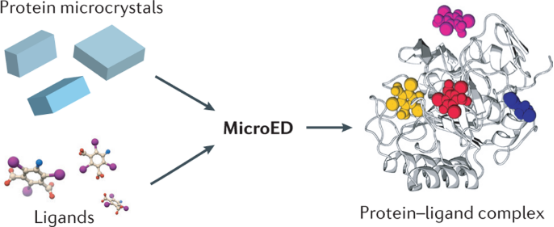
Figure 12. MicroED's Potential for Studying Protein-Ligand Interactions [12]
FAQ
Q1: What are the differences between MicroED and X-ray crystal diffraction?
MicroED requires smaller crystals. When crystallization experiments produce crystals thinner than ~400 nm, MicroED is an ideal choice, whereas larger and thicker crystals can be studied using traditional X-ray crystallography. Another difference between X-ray and electron diffraction is that X-ray crystallography data is typically collected using 12 keV X-ray photons, which have a highly curved Ewald sphere and larger scattering angles. MicroED generally uses 200 kV electrons, whose wavelength is much shorter, meaning the structure of the Ewald sphere is almost flat.
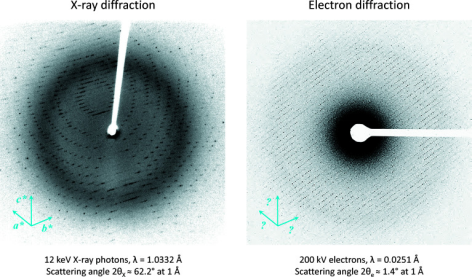
Figure 13. Differences Between X-ray and Electron Diffraction [6]
References
[1] Mackay JP, Landsberg MJ, Whitten AE, Bond CS. Whaddaya Know: A Guide to Uncertainty and Subjectivity in Structural Biology. Trends Biochem Sci. 2017 Feb;42(2):155-167. doi: 10.1016/j.tibs.2016.11.002. Epub 2017 Jan 12. PMID: 28089412.
[2] Rout MP, Sali A. Principles for Integrative Structural Biology Studies. Cell. 2019 May 30;177(6):1384-1403. doi: 10.1016/j.cell.2019.05.016. PMID: 31150619; PMCID: PMC6810593.
[3] Mu X, Gillman C, Nguyen C, Gonen T. An Overview of Microcrystal Electron Diffraction (MicroED). Annu Rev Biochem. 2021 Jun 20;90:431-450. doi: 10.1146/annurev-biochem-081720-020121. PMID: 34153215; PMCID: PMC9974886.
[4] Nguyen C, Gonen T. Beyond protein structure determination with MicroED. Curr Opin Struct Biol. 2020 Oct;64:51-58. doi: 10.1016/j.sbi.2020.05.018. Epub 2020 Jun 28. PMID: 32610218; PMCID: PMC7321661.
[5] Shi D, Nannenga BL, de la Cruz MJ, Liu J, Sawtelle S, Calero G, Reyes FE, Hattne J, Gonen T. The collection of MicroED data for macromolecular crystallography. Nat Protoc. 2016 May;11(5):895-904. doi: 10.1038/nprot.2016.046. Epub 2016 Apr 14. PMID: 27077331; PMCID: PMC5357465.
[6] Clabbers MTB, Shiriaeva A, Gonen T. MicroED: conception, practice and future opportunities. IUCrJ. 2022 Jan 18;9(Pt 2):169-179. doi: 10.1107/S2052252521013063. PMID: 35371502; PMCID: PMC8895021.
[7] Haymaker A, Bardin AA, Gonen T, Martynowycz MW, Nannenga BL. Structure determination of a DNA crystal by MicroED. Structure. 2023 Jul 24:S0969-2126(23)00248-4. doi: 10.1016/j.str.2023.07.005. Epub ahead of print. PMID: 37541248.
[8] Karothu DP, Alhaddad Z, Göb CR, Schürmann CJ, Bücker R, Naumov P. The Elusive Structure of Levocetirizine Dihydrochloride Determined by Electron Diffraction. Angew Chem Int Ed Engl. 2023 Jun 26;62(26):e202303761. doi: 10.1002/anie.202303761. Epub 2023 May 17. PMID: 37071841.
[9] Martynowycz MW, Shiriaeva A, Ge X, Hattne J, Nannenga BL, Cherezov V, Gonen T. MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP. Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2106041118. doi: 10.1073/pnas.2106041118. PMID: 34462357; PMCID: PMC8433539.
[10] Jones CG, Asay M, Kim LJ, Kleinsasser JF, Saha A, Fulton TJ, Berkley KR, Cascio D, Malyutin AG, Conley MP, Stoltz BM, Lavallo V, Rodríguez JA, Nelson HM. Characterization of Reactive Organometallic Species via MicroED. ACS Cent Sci. 2019 Sep 25;5(9):1507-1513. doi: 10.1021/acscentsci.9b00403. Epub 2019 Sep 6. PMID: 31572777; PMCID: PMC6764211.
[11] Danelius E, Patel K, Gonzalez B, Gonen T. MicroED in drug discovery. Curr Opin Struct Biol. 2023 Apr;79:102549. doi: 10.1016/j.sbi.2023.102549. Epub 2023 Feb 21. PMID: 36821888; PMCID: PMC10023408.
[12] Clark LJ, Bu G, Nannenga BL, Gonen T. MicroED for the study of protein-ligand interactions and the potential for drug discovery. Nat Rev Chem. 2021 Dec;5(12):853-858. doi: 10.1038/s41570-021-00332-y. Epub 2021 Oct 27. PMID: 37117388.
How to order?







