Native MS Service
- Structural analysis of proteins, nucleic acids, and sugars: For example, protein folding, secondary structure, glycosylation sites, etc.
- Interaction studies of large biomolecules and complexes: For example, protein-protein, protein-nucleic acid, protein-drug interactions, etc.
- Applications in drug development: Evaluate the binding performance of drugs to target proteins using native MS analysis technology.
- The purity of large biomolecules and complexes should be greater than 90%.
- Provide adequate sample quantity, usually a few micrograms to a few milligrams are needed.
- Please dissolve the sample in an appropriate non-denaturing solution, such as water or a buffer with a small amount of salt.
Native MS analysis is a spectrometry technique with an added ion source that can analyze large biomolecules and complexes under non-denaturing conditions. In native MS analysis, the sample is analyzed under conditions that are similar to the conditions inside the organism, thus preserving the native state of the sample. This technique has become an important tool for understanding the structure of proteins, nucleic acids, and sugars and their roles in complex biological processes.
Native MS analysis relies on the electrospray ionization (ESI) source to introduce ions from the solution of large biomolecules and complexes into the mass spectrometer. In ESI, the sample solution forms small droplets through a tiny nozzle, these droplets evaporate and ionize under high voltage, and then enter the mass spectrometer for detection.
Services at MtoZ Biolabs
MtoZ Biolabs has established a non-denatured protein profiling platform utilizing high-resolution liquid chromatography-mass spectrometry. This platform is capable of analyzing the structure, topology, and assembly of proteins and protein complexes that preserve their tertiary and quaternary structures in states closely resembling their natural conditions. Additionally, the platform facilitates the detailed characterization of monoclonal antibody/ADCs, PEG-modified proteins, oligomeric proteins, glycans, and assembled proteins.
1. Sample preparation: Dissolve the large biomolecule or complex in an appropriate non-denaturing solution.
2. Sample electrospray: Introduce the sample solution into the mass spectrometer through ESI.
3. Mass spectrometry data collection: Collect data using specific mass spectrometry techniques (e.g., static, dynamic, selective ion detection, etc.).
4. Data processing and analysis: Use specialized software to analyze and interpret the generated mass spectrometry data to draw conclusions.
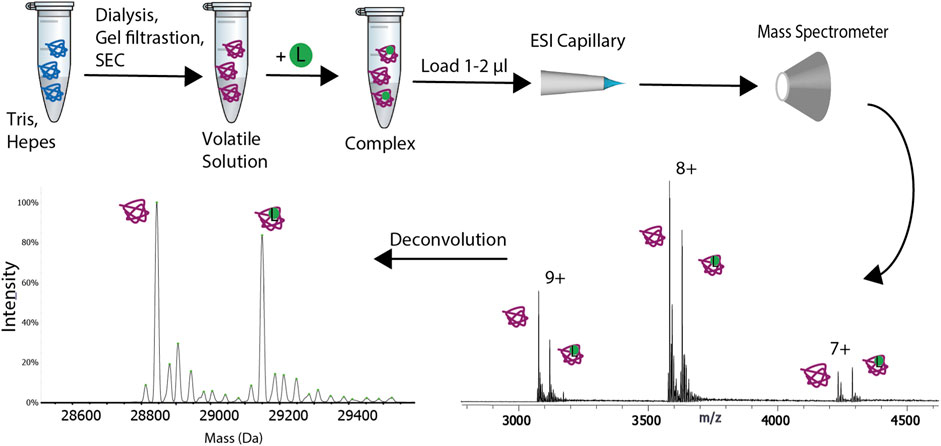
Gavriilidou, AFM. et al. Front Mol Biosci. 2022.
Figure1. Schematic Representation of the Experimental Procedure for Native Mass Spectrometry
Applications
Sample Submission Requirements
With its powerful technology, equipment, and talent advantages, MtoZ Biolabs provides professional native MS analysis services for customers. If you have any questions or specific needs, please feel free to contact us, and we will help you achieve your goals.
How to order?







