Native MS Analysis Service
Native MS analysis (Native mass spectrometry analysis) is an advanced mass spectrometry (MS) technique used to study the structure and function of proteins and their complexes under near-physiological conditions. Unlike traditional denaturing MS, native MS allows for precise analysis of the composition, structure, and interactions of protein complexes while preserving their native conformation and stability. Native MS analysis service offers unique advantages in studying protein-protein, protein-ligand, and protein-nucleic acid interactions, making it widely applicable in structural biology, drug discovery, and biomarker identification. Leveraging MS and chromatography platforms, MtoZ Biolabs provides native MS analysis service to reveal the molecular weight, binding states, conformational changes, and interactions of proteins in their native state, enabling researchers to gain deeper insights into biomolecular structure and function.
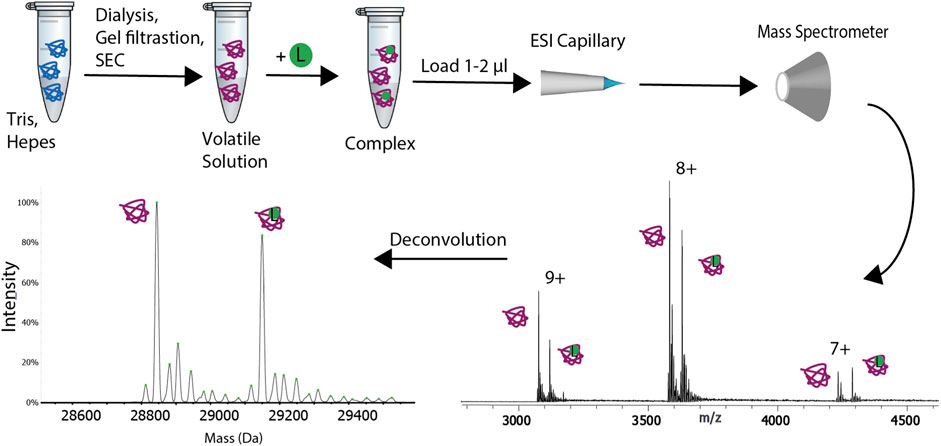
Gavriilidou AFM. et al. Front. Mol. Biosci. 2022.
Technical Principles
Native MS analysis dissolves proteins or protein complexes in a buffer solution that closely mimics physiological conditions to maintain their natural conformation and biological activity. Soft ionization techniques, most commonly electrospray ionization (ESI), are then used to introduce charged molecular ions into the MS system. ESI efficiently transfers samples from the liquid phase to the gas phase while preserving the native state of proteins and their complexes. During MS analysis, molecules remain in their native conformation and are separated and detected based on their mass-to-charge ratio (m/z).
Sample Submission Suggestions
Sample Types
We accept various types of samples, including but not limited to proteins, protein complexes, monoclonal antibodies, and post-translationally modified proteins.
Sample Quantity
A minimum of 50 μg is required, with the specific amount adjustable based on project requirements. Please contact us for consultation before sample submission.
Applications
Protein-Ligand Interaction Studies
Reveal the binding mechanisms between proteins and small-molecule drugs or natural ligands, providing structural insights for drug design.
Protein Complex Analysis
Analyze the composition, binding sites, and conformational changes of protein complexes, uncovering their functional mechanisms in cellular processes.
Post-Translational Modification (PTM) Studies
Investigate the effects of phosphorylation, glycosylation, and other PTM on protein structure and function.
Monoclonal Antibody Characterization
Identify antibody integrity, light-to-heavy chain ratios, and antigen-binding stability.
Biomacromolecule Drug Development
Evaluate the structural integrity, purity, and stability of biopharmaceuticals to ensure drug quality and efficacy.
FAQ
Q: How to prepare samples to maintain the native conformation of proteins or protein complexes and reduce non-specific binding or dissociation?
The core elements of sample preparation include buffer selection, ionic strength control, pH regulation and sample concentration management.
1. Buffer Selection
The most commonly used buffer is Ammonium Acetate (NH₄OAc) because it volatilizes quickly during non-denaturing ESI, minimizing interference with MS signals. For certain protein complexes, small amounts of glycerol or Guanidine HCl may be added to maintain protein solubility. However, the concentrations of these additives must be strictly controlled to avoid disrupting the native protein conformation.
2. Ionic Strength Control
Excessive ionic strength can cause protein aggregation or dissociation of protein complexes. Generally, it is recommended to keep the ionic strength of the buffer in the range of 50–200 mM, striking a balance between solubility and the stability of the native conformation.
3. pH Regulation
The isoelectric point (pI) of a protein is a key factor in determining its stability. The pH of the buffer should be close to the protein's physiological pH, typically within the range of pH 6.5–8.5, to prevent protein denaturation or aggregation caused by excessively acidic or alkaline conditions.
4. Sample Concentration Management
Overly high sample concentrations may lead to protein aggregation, while excessively low concentrations can reduce ESI efficiency. Typically, a sample concentration of 1–10 μM is recommended, but the specific concentration should be optimized based on the properties of the target protein.
5. Reduction of Non-Specific Binding
Adding a small amount of detergents (e.g., low concentrations of CHAPS or Tween-20) can effectively reduce non-specific interactions. Additionally, optimizing ESI parameters—such as voltage, spray gas flow rate, and temperature—can further minimize non-specific dissociation.
In Native MS analysis, obtaining accurate and reliable MS data depends on fully optimized sample preparation. Custom optimization is required for each protein or complex, with the primary goal of ensuring that the sample enters the mass spectrometer in its native conformation while minimizing structural damage during ionization.
Case Study
Native MS analysis was used to analyze the human pyruvate dehydrogenase complex (PDHc) and, for the first time, discovered the presence of (E2₁-E3BP₂)-type heterotrimers. Higher oligomers, namely dimers and trimers of the (E2₁-E3BP₂) heterotrimers, were also detected. This analysis provided new insights into the structure and function of PDHc, highlighting the power of Native MS in unraveling complex biological assemblies.
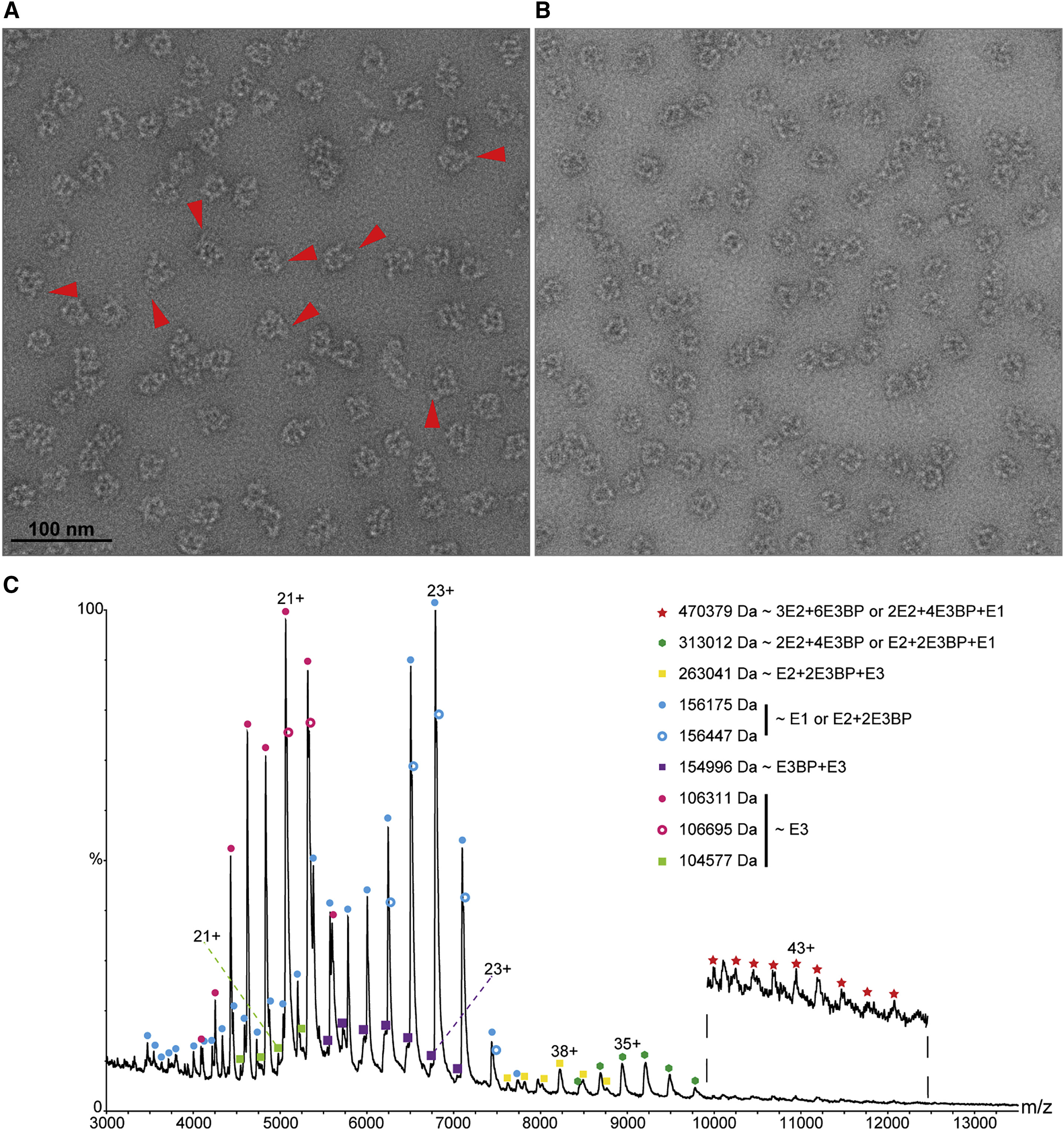
Prajapati S. et al. Structure. 2019.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
How to order?







