Peptide Mass Fingerprinting Analysis Service
Peptide mass fingerprinting (PMF), also termed protein fingerprinting, is a high-throughput analytical technique developed in 1933 for protein identification. It involves the cleavage of an unknown target protein into smaller peptides by endopeptidases, followed by the precise measurement of these peptides' absolute mass using a mass spectrometer to generate a peptide peak list. This list is then compared against a theoretical peptide mass database using bioinformatics tools. This database, which contains translated proteins from all known genomes, also predicts peptide masses post-hydrolysis, facilitating the identification process through statistical comparison of experimental peptide masses with theoretical values.
Analysis Workflow
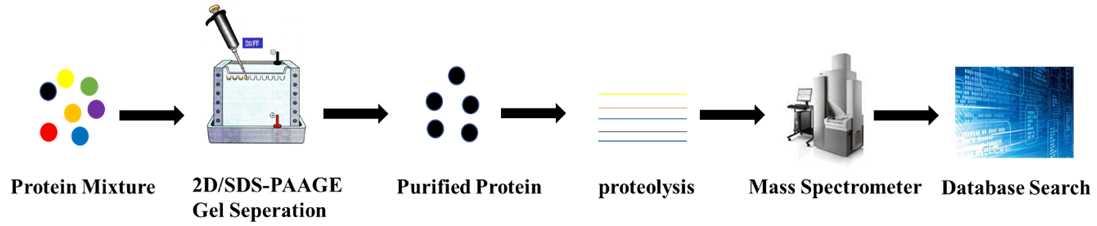
Figure 1. PMF Analytical Process
Proteins isolated from biological tissues or cells are separated via 2D or SDS-PAGE and analyzed using imaging techniques. Proteins showing significant differences are selected for further analysis, which involves gel extraction and enzymatic digestion to yield peptides that characterize the protein. The peptide mixture is then analyzed through mass spectrometry to obtain a detailed fingerprint spectrum, which aids in molecular weight determination and facilitates database matching.
Applications
PMF has gained widespread application across various domains, including drug authenticity verification, food quality control, and disease diagnosis, due to its high sensitivity, robustness, and minimal sample requirements. Recognized by the World Health Organization, it serves as a critical technique for material identification and quality control.
Service Advantages
1. Protein Identification Relies on Peptide Mass rather than Sequencing
2. Classical Method Employing First-Stage Mass Spectrometry for Rapid Protein Identification
Limitations
1. The Sequence of the Target Protein Must be Present in the Database
2. Primarily Applicable to Single Protein Analysis, the Presence of Mixed Proteins Complicates the Analysis
3. Similar Peptide Masses Increase the Challenge of Accurate Matching
MALDI-TOF PMF
Rapid and precise protein identification is essential in proteomics. PMF employs matrix-assisted laser desorption/ionization (MALDI) coupled with time-of-flight (TOF) mass spectrometry, making it a prevalent choice.
MALDI uses a laser to irradiate the eutectic film formed by the sample and substrate. The matrix absorbs energy from the laser and transfers it to the biomolecules. During the ionization process, protons are transferred to or from biomolecules, causing ionization. Thus, MALDI is a soft ionization technique, suitable for the determination of mixtures and biological macromolecules. The principle of TOF is that ions are accelerated through a flight tube under the influence of an electric field, and ions are detected according to the time it takes to fly to the detector. MALDI-TOF-MS has high sensitivity, precision, and resolution. It plays a crucial role in sample analysis in life sciences and many other fields.
MtoZ Biolabs provides specialized protein identification services utilizing peptide fingerprinting PMF technology, and welcomes your inquiries.
Deliverables
In the technical report, MtoZ Biolabs will provide you with a detailed technical report, including:
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Mass Spectrometry Images
4. Raw Data
5. Results of Protein Molecular Weight and Purity Analysis
How to order?







