Principal Component Analysis (PCA) Service
Principal component analysis (PCA) is the linear recombination of all metabolites identified to form a set of new comprehensive variables, and select 2-3 variables, reflecting as much information of the original variables as possible, based on the analyzed problem to achieve the purpose of dimension reduction. At the same time, PCA of metabolites can also reflect the variability between and within groups overall. The total sample PCA analysis uses the PCA method to observe the overall distribution trend between all group samples, identify potential outlier samples, and consider various factors (number of samples, preciousness of samples, dispersion degree) to decide whether to remove outlier points. The PCA score chart for all samples is shown below (PCA score chart for pairwise analysis of samples).
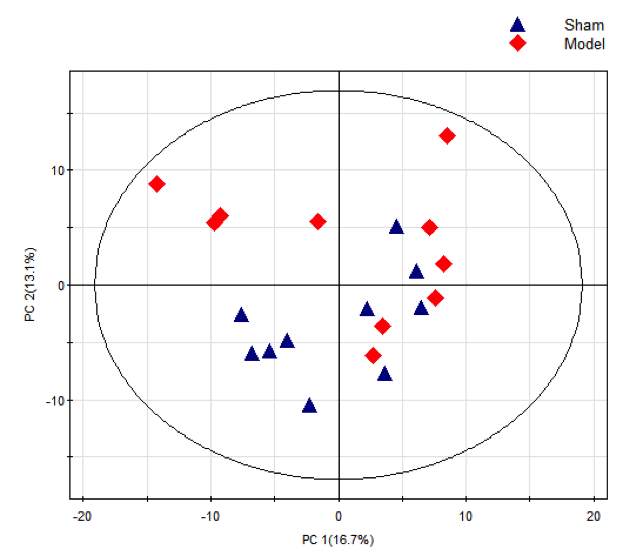
Figure 1. PCA Score Chart
MtoZ Biolabs uses XCMS software to extract ion peaks of metabolites. The peaks extracted from 25 samples and QC samples, after normalization, undergo PCA analysis. As shown in the chart, QC samples (grey) are closely clustered together, indicating good stability of the instrument analysis system in this experiment, reliable experimental data. The metabolic spectrum differences obtained in the experiment can reflect the biological differences between samples.
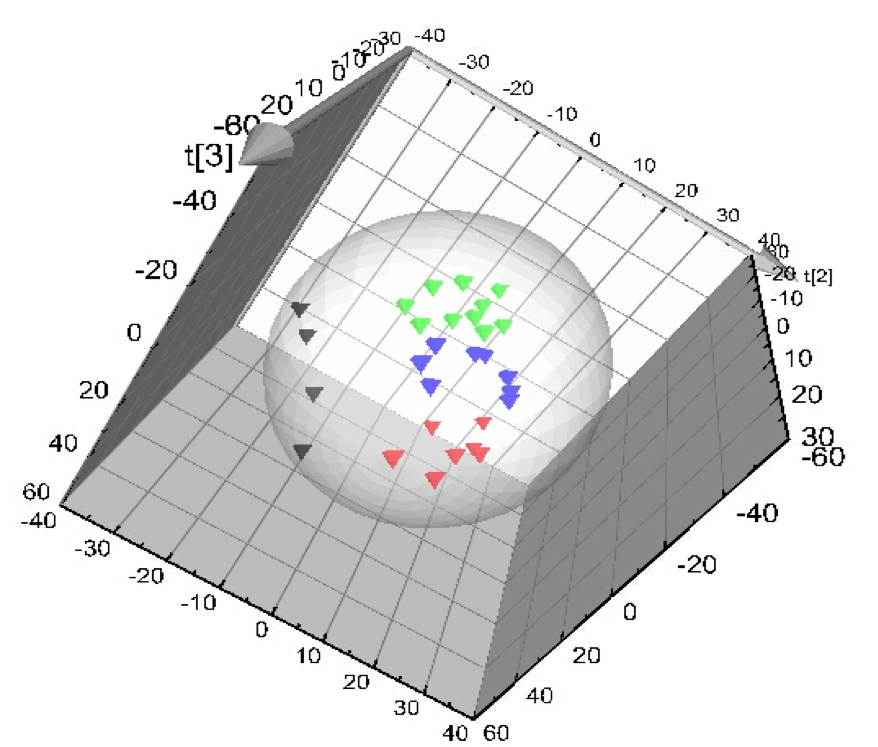
Figure 2. Total Sample PCA Score Chart
How to order?







