Molecular Glue Screening Service | PROTAC Service
Proteins play a pivotal role in the regulation, function, and structure of various human organs and tissues. The regulation of the synthesis and degradation of proteins is crucial for cell functionality. Protein turnover can exist in three states: equilibrium, pool expansion, and pool contraction. At equilibrium, the rate of protein synthesis matches that of degradation, leading to unchanged transcriptome and metabolome profiles. Expansion of the protein pool occurs either through an increase in synthesis or a decrease in degradation, which necessitates adjustments in the transcriptome or metabolome accordingly. A contraction in the protein pool results from a decrease in synthesis (affecting the transcriptome) or an increase in degradation (affecting the metabolome). Changes in a protein's structure, stereochemistry, localization, function, or equilibrium can disrupt protein homeostasis, potentially leading to cellular dysfunction, death, or severe diseases.
The degradation of proteins is essential for cell processes and the maintenance of cellular protein equilibrium, involving mechanisms like cell cycle regulation, inflammatory transcription gene responses, apoptosis, and DNA repair. In eukaryotic cells, two principal pathways mediate protein degradation: the lysosomal and the ubiquitin-proteasome pathways. The lysosomal pathway degrades both extracellular and intracellular materials (such as proteins, nucleic acids, and polysaccharides). Through endocytosis, extracellular substances and membrane components (e.g., receptors or channels) are internalized and degraded in acidic endosomes/lysosomes, contributing to membrane repair, immune responses, bone resorption, and pathogen elimination. Lysosomes also degrade damaged intracellular proteins or organelles via autophagy, which involves the encapsulation of damaged proteins by autophagosomes. Regulating intracellular substance levels through lysosomes is vital for sustaining normal metabolic activities and microbial infection resistance. The proteasome system represents the second major degradation pathway, wherein the 26S proteasome complex degrades proteins into peptides. This system comprises the proteasome, various ubiquitin ligases, and ubiquitinating/deubiquitinating enzymes. Ubiquitin is a highly conserved regulatory protein in all forms of life. It consists of 76 amino acids and has a molecular weight of 8.5 kDa. It is linked to target degradation proteins through a multi-step process called ubiquitination. Ubiquitination involves the action of three enzymes, mediating the covalent modification of proteins. In the first step, E1 (ubiquitin-activating enzyme) binds to the ubiquitin molecule at its active site. Next, ubiquitin is transferred to the active site of E2 (ubiquitin-conjugating enzyme). In the third step, E3 (ubiquitin ligase) identifies the substrate protein to be degraded and catalyzes the transfer of ubiquitin from the E2 protein to the substrate.
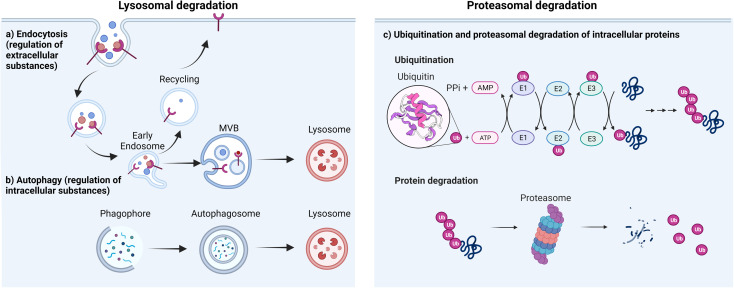
Figure 1. Main Pathways of Protein Degradation [1]
The misfolding, overexpression, and aggregation of proteins are implicated in the pathogenesis of neurodegenerative diseases, cancers, and autoimmune disorders. Targeting these proteins with small molecule drugs represents a promising therapeutic strategy. Yet, certain proteins, like scaffold proteins, transcription factors, and non-enzymatic proteins, are considered "undruggable" due to the infeasibility of targeting them with small molecules. Moreover, the occupancy-driven pharmacodynamics of some small molecule drugs, such as kinase inhibitors, can lead to off-target effects and necessitate higher plasma concentrations for efficacy. The concept of inducing protein degradation emerges as an innovative pharmacological strategy, encompassing technologies like proteolysis targeting chimeras (PROTACs), hydrophobic tags (HyTs), selective estrogen receptor degraders (SERDs), lysosome targeting chimeras (LYTACs), autophagy-targeting chimeras (AUTACs), and molecular glues.
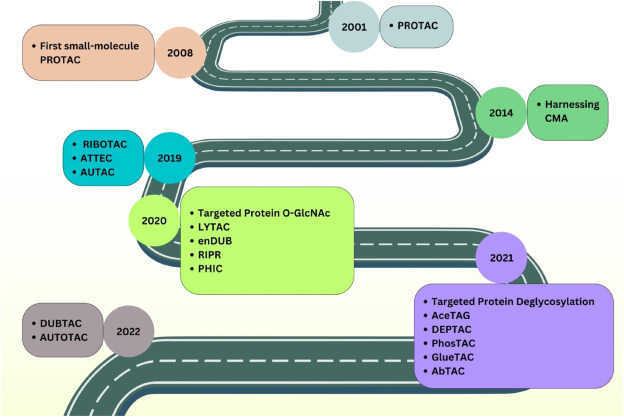
Figure 2. Development History of Targeted Protein Modification (TPM) Technology [2]
Clinically tested targeted protein degradation therapies are categorized into immunomodulatory drugs (IMiDs), selective estrogen receptor degraders (SERDs), and PROTACs, based on their design and mechanism. Though all operate through the ubiquitin-proteasome system (UPS), their ubiquitination efficiencies vary, leading to distinct clinical applications. IMiDs, functioning as "molecular glue," interact with cereblon (CRBN) to recruit various proteins for degradation. PROTACs, with dual ligands, bind target proteins and E3 ubiquitin ligase to facilitate ubiquitin transfer and subsequent degradation of the protein. Unlike PROTACs, SERDs induce conformational changes in estrogen receptor α (ERα), making it recognizable for ubiquitination and proteolysis. These distinctions in ubiquitination and degradation mechanisms underpin the unique clinical implications and outcomes of these therapies.
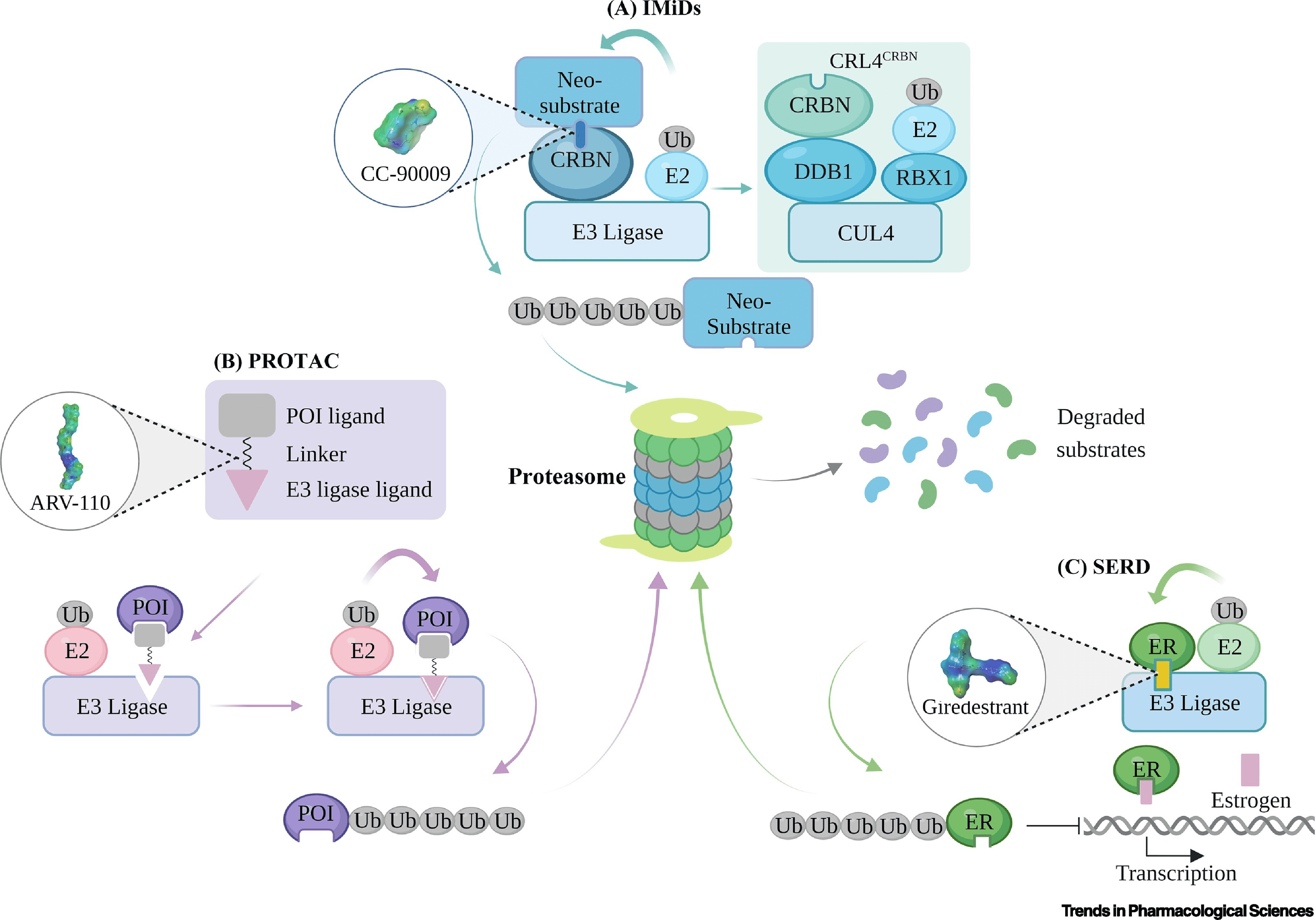
Figure 3. Schematic of Targeted Protein Degradation Mechanisms [3]
The distinction between small molecule inhibitors and targeted protein degraders lies in their mechanisms of action: inhibitors block the function of specific targets, while degraders eliminate the targets of interest. Compared to small molecule inhibitors, PROTAC degraders offer several advantages: the degradation of a protein by PROTAC affects both its enzymatic and non-enzymatic functions, whereas small molecule drugs do not impact non-enzymatic functions; PROTACs exhibit greater sensitivity to drug-resistant targets; PROTACs can potentially target molecules that are otherwise considered undruggable; and PROTACs are more potent at nanomolar concentrations, whereas traditional drugs require higher concentrations to achieve clinical efficacy due to off-target effects. Consequently, PROTACs and similar degraders are deemed effective for cancer treatment. Currently, more than 85 targeted protein degraders are in clinical or pre-clinical/development stages. Various commercial and public investors have invested in this area of research. Four PROTAC technology companies have gone public, achieving a cumulative market value of 11 billion USD.
MtoZ Biolabs possesses a comprehensive pre-clinical platform for the development of targeted protein degradation drugs, offering services in PROTAC target selection, molecular design, molecular modification and synthesis, and screening validation. The team is capable of characterizing the physicochemical properties of PROTAC ternary complexes through a variety of techniques, such as fluorescence polarization or time-resolved fluorescence resonance energy transfer, Western blot, and ubiquitination detection. This allows for the determination of affinity for target proteins and the measurement of ubiquitination degradation efficiency, enabling the selection of PROTACs with the best target degradation efficiency. Furthermore, MtoZ Biolabs can conduct multi-level testing, including at the molecular, cellular, and animal levels. Mtoz Biolabs provides a one-stop solution for PROTAC resource libraries, PROTAC synthesis, pharmacological screening models, and screening data analysis, addressing all aspects of PROTAC design, synthesis, and screening.
References
[1] Shen F, Dassama LMK. Opportunities and challenges of protein-based targeted protein degradation. Chem Sci. 2023 Jul 3;14(32):8433-8447. doi: 10.1039/d3sc02361c. PMID: 37592990; PMCID: PMC10430753.
[2] Amirian R, Azadi Badrbani M, Izadi Z, Samadian H, Bahrami G, Sarvari S, Abdolmaleki S, Nabavi SM, Derakhshankhah H, Jaymand M. Targeted protein modification as a paradigm shift in drug discovery. Eur J Med Chem. 2023 Nov 15;260:115765. doi: 10.1016/j.ejmech.2023.115765. Epub 2023 Aug 29. PMID: 37659194.
[3] Fang Y, Wang S, Han S, Zhao Y, Yu C, Liu H, Li N. Targeted protein degrader development for cancer: advances, challenges, and opportunities. Trends Pharmacol Sci. 2023 May;44(5):303-317. doi: 10.1016/j.tips.2023.03.003. PMID: 37059054.
How to order?







