Protein Interaction Network Analysis Service
STRING is a database that records predicted and experimentally validated protein-protein interactions (PPIs) across multiple species, including direct physical interactions and indirect functional associations. By integrating the results of differential expression analysis and the interaction pairs recorded in the database, a differential expression protein interaction network is constructed. During the analysis, differentially expressed proteins are mapped to the STRING database to obtain information on their interaction relationships. As the STRING database includes experimental data, results from text mining of PubMed abstracts, and aggregated data from other databases, as well as predictions made using bioinformatics methods, we select interaction pairs with a combined score greater than 0.4 (Medium) from the search results and use appropriate bioinformatics analysis software to visualize the interaction results. The results are shown in the below:
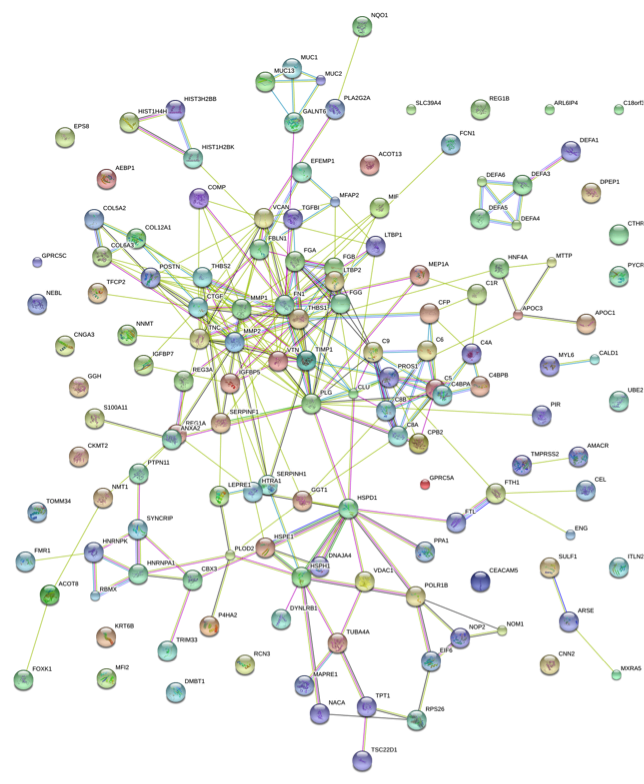 Figure 1. Evidence of Differential Expressed Protein Interactions from Different Sources (Evidence)
Figure 1. Evidence of Differential Expressed Protein Interactions from Different Sources (Evidence)

Evidence of Differential Expressed Protein Interactions from Different Sources (Evidence) 2
 Figure 2. Graph of Differential Expressed Protein Interaction Strengths (Confidence)
Figure 2. Graph of Differential Expressed Protein Interaction Strengths (Confidence)
Note: Circles (nodes) represent proteins; Different colors indicate different proteins, and the circles contain the three-dimensional structure of the proteins. Lines indicate the interactions between proteins. Thicker lines indicate stronger interaction relationships. Dark lines indicate relationships in the evidence view circle but no relationships in the actions diagram.
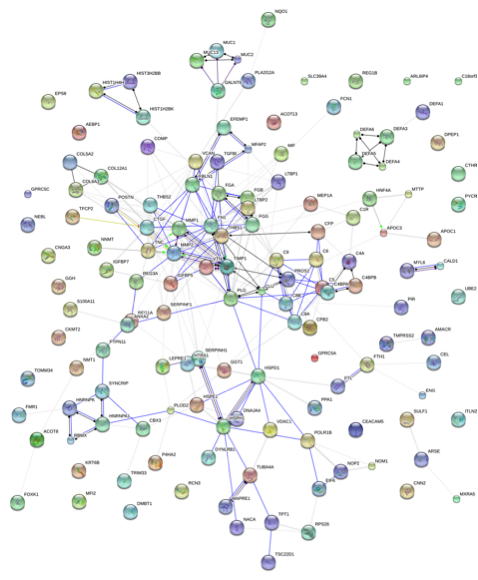 Figure 3: Different Action Modes between Differentially Expressed Proteins (Actions)
Figure 3: Different Action Modes between Differentially Expressed Proteins (Actions)

Different Action Modes between Differentially Expressed Proteins (Actions) 2
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







