Protein Sequencing
- PVDF membranes
- Gel bands or gel spots
- Protein solutions (preferably in volatile or buffer-free conditions)
The primary structure of a protein, its linear amino acid sequence, forms the basis for its folding, function, and interactions. Determining this sequence is critical for accurate protein characterization, antibody development, biosimilar verification, and intellectual property protection.
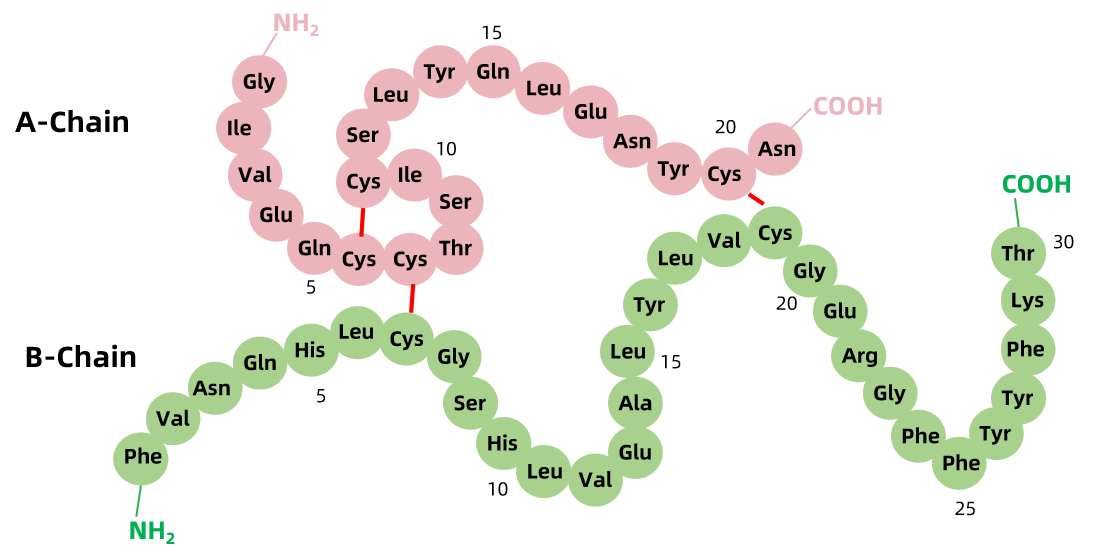
Figure 1. Protein Primary Structure
MtoZ Biolabs offers comprehensive Protein Sequencing Service powered by high-resolution mass spectrometry (MS) and Edman degradation technologies. Our platform supports full-length sequencing, terminal sequence confirmation, de novo sequencing of novel proteins, and antibody sequence elucidation. The Protein Sequencing Service helps researchers and biopharmaceutical companies validate expression products, characterize therapeutic antibodies, and uncover unknown or modified protein variants with high confidence.
Services at MtoZ Biolabs
MtoZ Biolabs provides a full spectrum of Protein Sequencing Service to address diverse research and development needs:
Determines the starting amino acids using Edman degradation or LC-MS/MS. This method is widely used to confirm the expression start site of recombinant proteins or define the N-terminus of antibody chains.
Combines protease digestion and tandem mass spectrometry to identify C-terminal residues. Ideal for validating truncations, fusion tags, or post-translational modifications at the protein’s C-terminus.
· Protein Full-length Sequencing
Employs a multi-enzyme digestion strategy and high-resolution MS to reconstruct the complete amino acid sequence. Suitable for native proteins, engineered constructs, and protein domains.
Reconstructs protein sequences directly from MS/MS spectra without relying on database searches. Especially beneficial for novel proteins, non-model organisms, or natural products.
Used to verify synthetic peptide sequences, detect impurities, and assess peptide integrity or modifications such as disulfide bonds or cyclization.
Provides full heavy and light chain sequence analysis from hybridoma or recombinant antibodies. CDR (complementarity-determining region) annotation supports applications in humanization, affinity maturation, and patent filing.
Technologies and Workflow
1. Mass Spectrometry-Based Sequencing
MtoZ Biolabs leverages Thermo Scientific™ Orbitrap Fusion™ Lumos™ and Q Exactive HF mass spectrometers equipped with both HCD and ETD fragmentation to provide high-resolution, high-accuracy protein sequencing.
We utilize a six-protease digestion strategy that includes Trypsin, Chymotrypsin, Lys-C, Lys-N, Glu-C, and Asp-N to generate overlapping peptides, enabling greater than 95% sequence coverage, even for complex or modified proteins.

Figure 2. Protein Sequencing by Mass Spectrometry
2. Edman Degradation
As a classical sequencing method, Edman degradation accurately identifies up to 30 amino acids from the free N-terminus. While not suitable for blocked or modified termini, it remains a valuable complement to MS-based approaches.
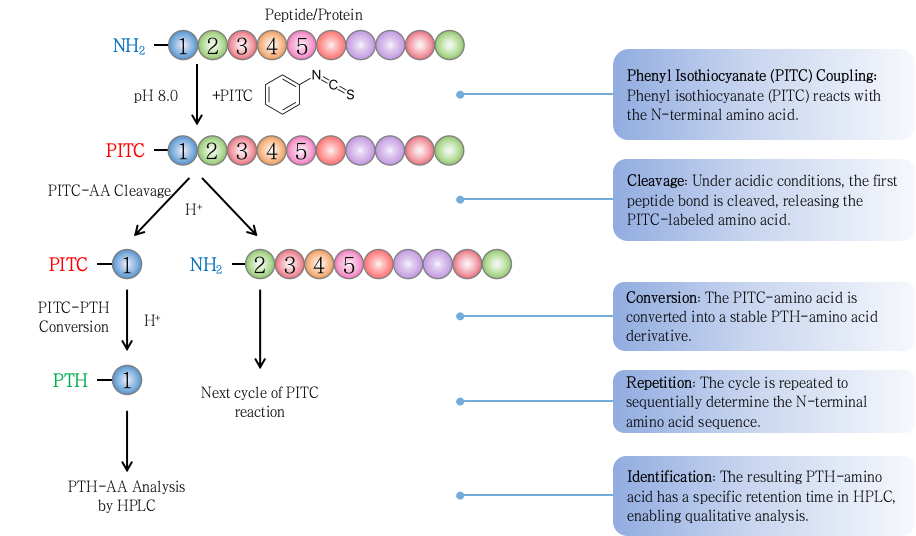
Figure 3. N-terminal Sequencing by Edman Degradation
Automated Workflow
Our protein sequencing pipeline is fully automated, covering everything from enzymatic digestion and MS data acquisition to spectrum interpretation and sequence assembly, ensuring high-throughput capacity, consistent data quality, and minimal human error.
Why Choose MtoZ Biolabs?
✅ High-Precision Sequencing
State-of-the-art instrumentation allows for precise amino acid resolution and confident annotation of post-translational modifications (e.g., phosphorylation, acetylation, glycosylation), essential for structural studies and mutation mapping.
✅ Broad Sample Compatibility
Our protocols support a wide range of proteins, including truncated fragments, chemically modified variants, native proteins, and low-abundance targets such as antibody fragments and peptide drugs.
✅ Streamlined, End-to-End Workflow
Standardized and automated processes ensure rapid turnaround, reproducible results, and consistent quality, making our services ideal for research, regulatory submissions, and publication needs.
✅ Maximal Sequence Coverage
Our six-enzyme digestion strategy ensures comprehensive and redundant peptide mapping, enabling accurate reconstruction of full-length protein sequences from microgram-level inputs.
✅ Comprehensive Deliverables
Clients receive:
Full-length amino acid sequences (FASTA format)
Annotated MS/MS spectra
Sequence coverage maps
Detected mutations and PTMs
Technical report for documentation and regulatory compliance
All results are compatible with bioinformatics tools such as Mascot and PEAKS, and are suitable for publication or intellectual property applications.
Sample Submission Suggestions
To maximize sequencing accuracy, samples should be as pure as possible. We recommend pre-treatment using SDS-PAGE, HPLC, or capillary electrophoresis, depending on sample type and analysis goals.
Accepted sample formats include:
Ship samples with appropriate temperature controls (ice packs or dry ice) to maintain protein integrity. If needed, MtoZ Biolabs provides comprehensive sample preparation services to streamline the entire workflow.
MtoZ Biolabs is committed to providing a one-stop Protein Sequencing Service for researchers and biotechnology companies. Feel free to contact us by phone or email, and we will provide professional consultation and support.
How to order?







