PTM Analysis Service
PTM analysis service is used to analyze post-translational modifications (such as phosphorylation, glycosylation, acetylation) of proteins, allowing the identification of PTM modification types and modification sites. Post-translational modifications (PTMs) refer to chemical modifications of amino acid residues that occur after protein translation, either through enzymatic or non-enzymatic processes. By altering protein structure, stability and activity, PTMs play a critical role in various cellular processes. Therefore, PTM analysis is extremely important in cell biology and related research fields. Based on chromatography and mass spectrometry technologies, MtoZ Biolabs provides PTM analysis service to help uncover the molecular mechanisms regulating protein function. We can identify multiple PTMs, including phosphorylation, glycosylation, ubiquitination, acetylation, methylation, disulfide bonds, and S-nitrosylation.
Laboratory C R. et al. Molecules. 2023.
Analysis Workflow
The primary steps for PTM analysis service of proteins include:
Sample Preparation and Protein Extraction: Extract total proteins from samples such as cells, tissues, or bodily fluids, then perform quantification and quality assessment.
Enzymatic Digestion and Peptide Preparation: Use specific enzymes (e.g., trypsin) to digest proteins into detectable peptide fragments.
PTM Peptide Enrichment: Enrich target modified peptides using specific strategies (such as antibody enrichment or TiO₂ enrichment).
LC Separation: Employ nano liquid chromatography (Nano-LC) for efficient separation of complex peptide mixtures.
MS/MS Detection: Use high-resolution mass spectrometers (e.g., Orbitrap Fusion Lumos) to accurately detect peptide fragments and PTM sites.
Data Analysis: Combine specialized databases and bioinformatics tools to identify PTMs, locate modification sites, and perform quantitative analysis.
Result Interpretation: Provide a detailed report.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced PTM mass spectrometry analysis platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all PTM mass spectrometry analysis data, providing clients with a comprehensive data report.
4. Offering Multiple PTM Analysis: MtoZ Biolabs employs high-resolution mass spectrometry platforms combined with Nano-LC to provide clients with the identification of PTM such as phosphorylation, glycosylation, ubiquitination, acetylation, methylation, disulfide bonds, and S-nitrosylation.
Applications
Protein PTM analysis is widely applied in various biological and medical fields. Below are some typical application areas:
1. Disease Mechanism Research
Many diseases are closely associated with abnormal PTMs. For example, dysphosphorylation in cancer signaling pathways. PTM analysis helps researchers understand the molecular mechanisms of disease onset and progression.
2. Drug Target Discovery and Drug Development
Identify disease-related PTM target proteins to aid in developing novel therapeutic agents. Assess the influence of drugs on specific PTMs to optimize drug design.
3. Cellular Signaling Pathway Analysis
Investigate PTMs of proteins in key signaling pathways to uncover mechanisms of signal transduction. Analyze the regulatory networks of PTMs between transcription factors and signaling pathways.
4. Bioengineering and Synthetic Biology
Optimize PTM patterns in recombinant proteins to enhance stability and functional activity of therapeutic proteins.
5. Environmental Stress Response
Examine how PTMs affect cell survival and adaptation under environmental stimuli.
FAQ
Q: In complex biological samples, the modification signals of specific PTM sites are often weak and easily masked by other strong signals. How can precise identification of these PTM sites be achieved?
To achieve precise identification of PTM sites, it is essential to enhance four key aspects through coordinated efforts: sample preparation, enrichment strategies, mass spectrometry parameter optimization, and data analysis.
1. Sample Preparation
Utilize high-quality sample preparation workflows to prevent protein degradation and PTM modification loss.
Ensure that non-specific modifications are avoided during extraction, digestion, and purification processes.
2. Enrichment Strategies
Employ specific enrichment techniques, such as using TiO₂ or IMAC (Immobilized Metal Affinity Chromatography) for phosphorylated peptides and specific antibodies for acetylated peptides.
For multiple PTM modifications, adopt dual enrichment strategies to progressively reduce background interference.
3. Mass Spectrometry Parameter Optimization
Choose appropriate mass spectrometry fragmentation modes (e.g., CID, HCD, ETD) to ensure effective cleavage of peptide fragments surrounding modification sites.
Optimize fragmentation energy parameters to prevent over-fragmentation or signal loss.
4. Data Analysis
Use high-quality databases (e.g., UniProt, PhosphoSitePlus).
Optimize mass spectrometry data search parameters to reduce the False Discovery Rate (FDR).
Utilize software tools (e.g., MaxQuant, Proteome Discoverer) for precise localization and statistical analysis of PTM sites.
Key Breakthroughs:
Combining multiple enrichment strategies with high-resolution mass spectrometry can significantly increase the coverage of PTM site identification.
Utilizing quantitative labeling techniques (e.g., TMT and iTRAQ) allows for the effective capture of low-abundance PTM signals in complex samples.
Case Study
PTM analysis discovered significant differences in the post-translational modification levels of different splice variants of the histone deacetylase HDAC7, particularly in phosphorylation modifications. This finding aids in revealing how alternative splicing of HDAC7 affects its interaction with 14-3-3 proteins.
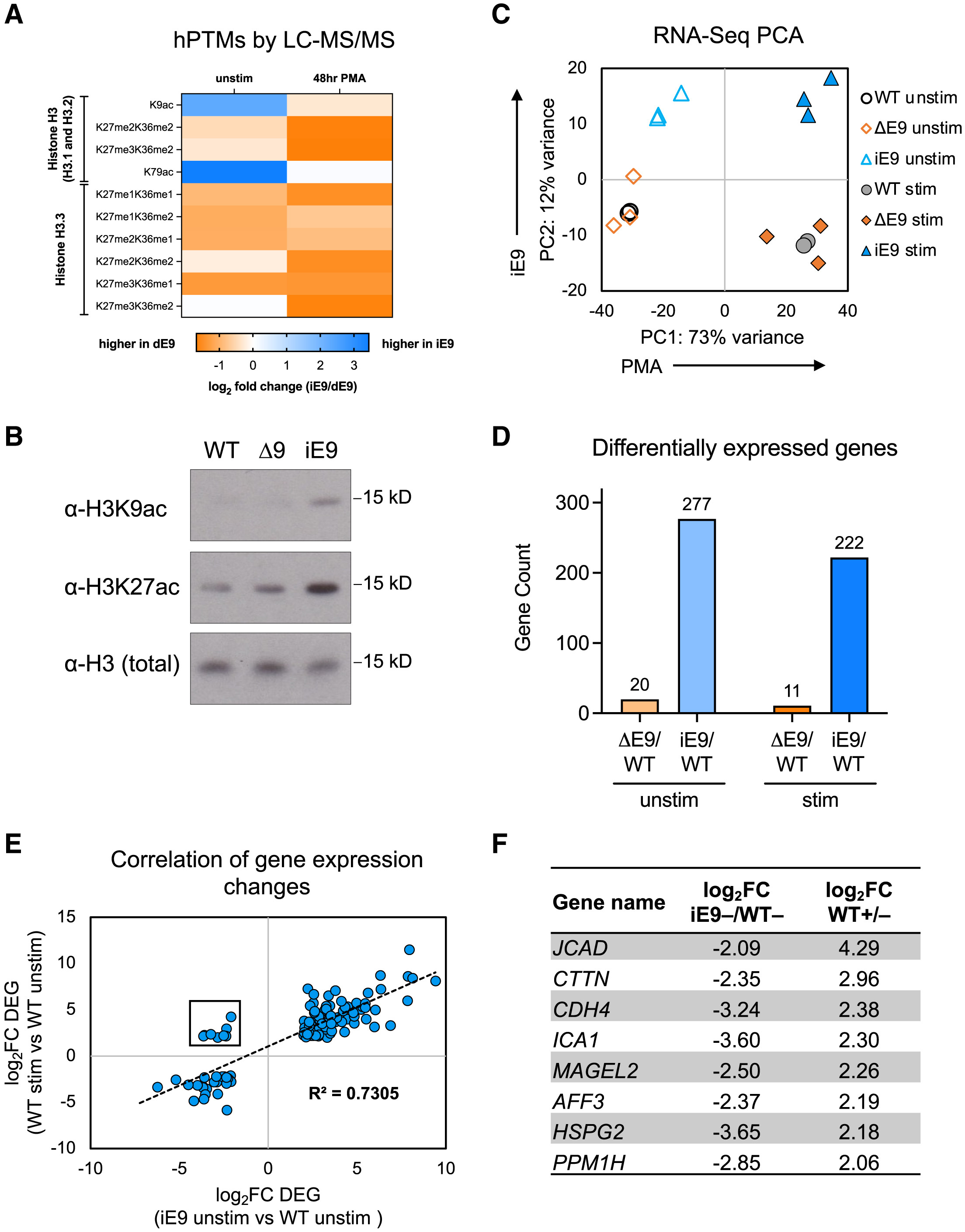
Agosto L M. et al. Cell reports. 2023.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Post-Translational Modification Functional Analysis Service
PRM-based Post-translational Modification Site Analysis Service
Histone Post-Translational Modification (PTM) Proteomics Service
How to order?







