PTM Mass Spectrometry Analysis Service
PTM mass spectrometry analysis service utilizes high-resolution mass spectrometers to identify and quantify post-translational modifications (PTMs) of proteins. Common PTMs include phosphorylation, acetylation, methylation, ubiquitination, and glycosylation, among others. These modifications can greatly influence the biological functions of proteins which are involved in a wide range of biological processes, such as signal transduction, cell cycle regulation, and gene expression regulation. Therefore, analyzing protein post-translational modifications helps in understanding the biological functions of proteins and elucidating related biological processes. Through High-Performance Liquid Chromatography–Tandem Mass Spectrometry (HPLC-MS/MS) technology, PTM mass spectrometry analysis can accurately localize protein post-translational modification sites, identify types of modifications, and perform quantitative analysis of modification levels. Leveraging the advantages of mass spectrometry technology, PTM mass spectrometry analysis service provided by MtoZ Biolabs can efficiently and accurately identify and quantify protein post-translational modifications, thereby supporting customers' related research work.
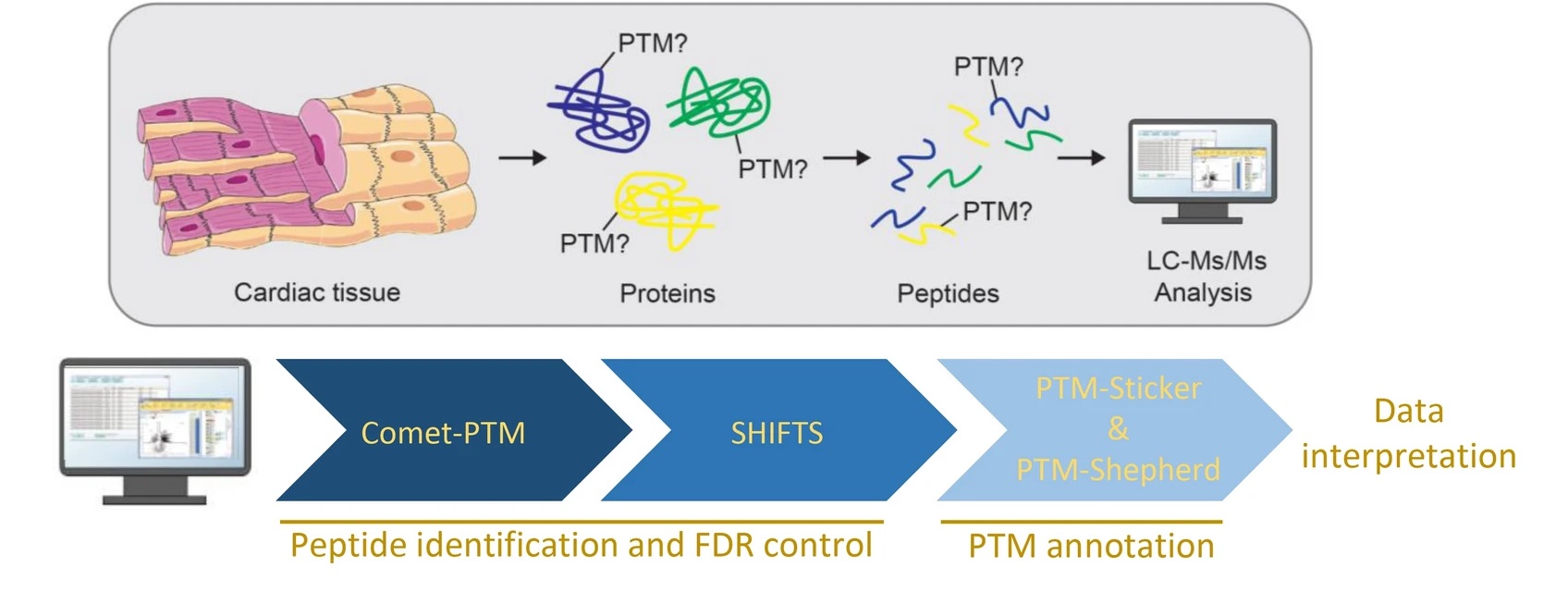
Bagwan N. et al. Scientific Reports. 2021.
Technical Principles
The core principle of PTM mass spectrometry analysis lies in the precise analysis of proteins and their modifications using mass spectrometers. Compared with unmodified proteins, PTM lead to an increase in specific molecular weights. Through mass spectrometry, a series of peptide molecular masses can be obtained. When proteins are not modified, both the sequence information and molecular weight of each peptide are fixed. However, once a PTM occurs—for instance, phosphorylation—some peptides exhibit an increase in molecular weight precisely corresponding to a phosphate group. It can then be inferred that these peptides are phosphorylated, which can be further confirmed by secondary mass spectrometry. By combining specific databases and bioinformatics tools, it is possible to pinpoint PTM modification sites and perform both qualitative and quantitative analyses of the PTM.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Using high-resolution mass spectrometry platforms combined with Nano-LC, MtoZ Biolabs provides clients with the identification of PTM such as phosphorylation, glycosylation, ubiquitination, acetylation, methylation, disulfide bonds, and S-nitrosylation.
Applications
PTM mass spectrometry analysis technology is widely applied in various life science fields, including but not limited to:
Drug Development and Precision Medicine
Through in-depth analysis of PTMs, potential biomarkers and drug targets can be discovered.
Cancer Research
Specific PTM patterns often exist in tumor cells. PTM mass spectrometry analysis can help uncover the molecular mechanisms associated with cancer.
Neuroscience
Protein PTMs in the nervous system are crucial for neural signal transmission and the development of neurodegenerative diseases.
Cell Biology and Developmental Biology
By studying the modification states of intracellular proteins, researchers can understand cell division, differentiation, and functional regulation.
FAQ
Q: How can multiple modification interferences in complex samples be effectively managed?
Complex samples often contain various types of PTMs, which may occur on the same protein or involve cross-modifications across multiple proteins, posing significant challenges for identification and quantification. The following strategies can be employed to handle multiple modification interferences in complex samples:
Step-by-step Enrichment Strategy
Utilize specific antibodies or affinity chromatography techniques to enrich particular modifications (e.g., phosphorylation, methylation, acetylation). By performing enrichment step by step, the signal intensity of target modifications is enhanced, thereby improving the sensitivity for detecting modification sites.
Multi-stage Mass Spectrometry Analysis
Implement multi-stage mass spectrometry strategies such as tandem mass spectrometry (MS/MS), high energy collisional dissociation (HCD), and electron transfer dissociation (ETD).
These techniques effectively distinguish the diversity of modifications. For instance, HCD mass spectrometry data can differentiate how different modifications affect the fragmentation patterns of precursor ions, allowing for the separation of overlapping modification signals.
High-throughput Data Analysis and Algorithms
Employ efficient mass spectrometry data analysis software capable of handling high-throughput and complex data. Utilize advanced algorithms to eliminate non-target modification signals, focusing on the identification and quantification of specific modifications.
Standardization and Quantitative Analysis
Incorporate stable isotope-labeled internal standards or standard samples. Use quantitative mass spectrometry techniques such as label-free quantification, TMT (Tandem Mass Tag), or iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) to enhance the quantitative accuracy of modifications, thereby avoiding quantification biases introduced by sample complexity.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
Using PTM mass spectrometry analysis to identify the PTMs of MYC in various experimental samples revealed that the ubiquitin ligase TRAF6 can ubiquitinate MYC at K148. The loss of TRAF6 and the corresponding increase in MYCK148 acetylation results in enhanced MYC-induced oncogenic function and development of AML.
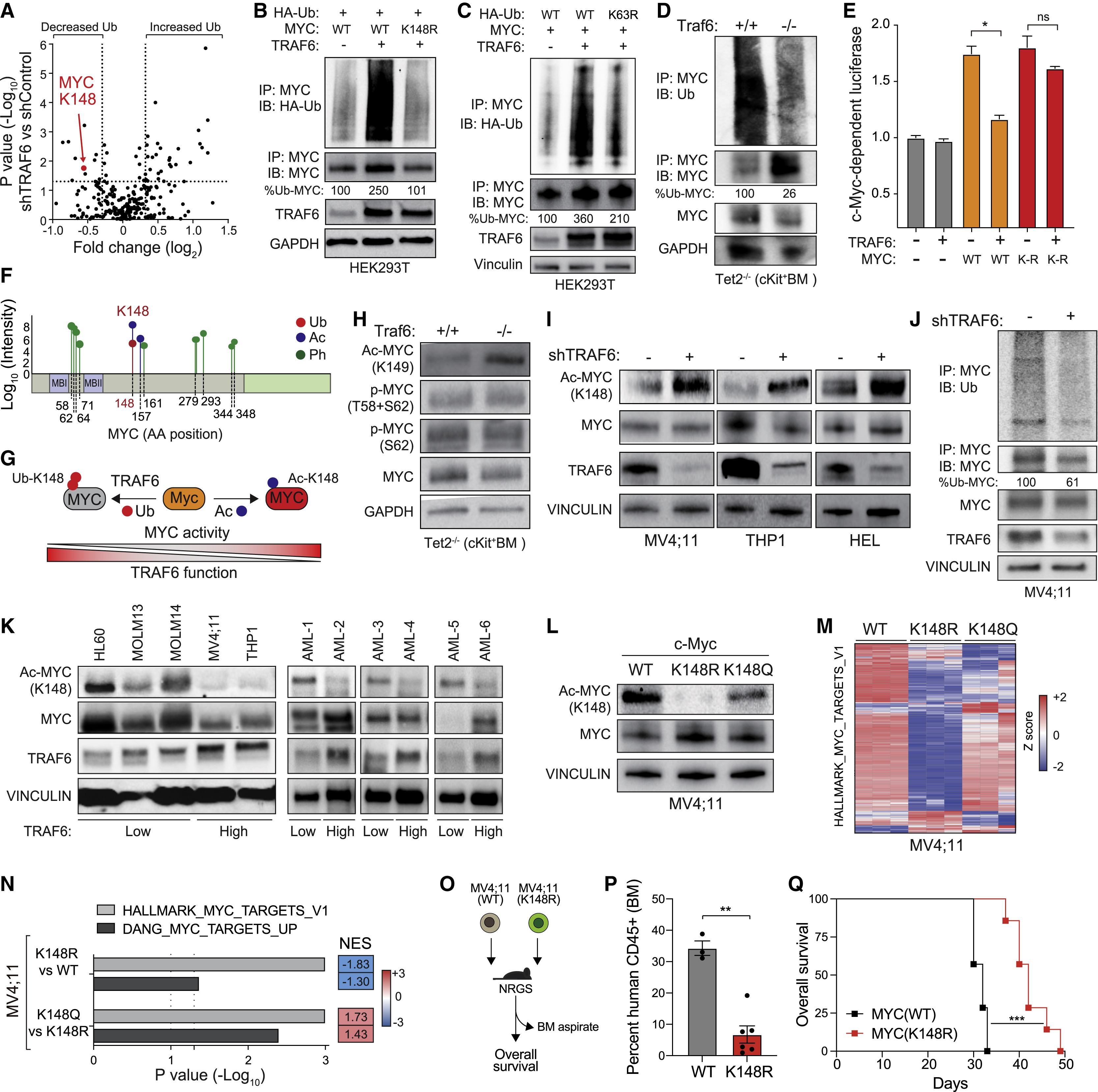
Muto T. et al. Cell stem cell. 2022.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
PRM-based Post-translational Modification Site Analysis Service
Histone Post-Translational Modification (PTM) Proteomics Service
How to order?







