Pull Down based Protein Analysis Service with Mass Spectrometry
In the biological activities of organisms, the function of proteins is achieved through interactions between proteins. Therefore, studying new protein interactions can provide new directions for investigating the potential functions of proteins. Among many protein interaction identification experiments, pull-down is commonly used for in vitro protein interaction testing, and helps discover new interacting proteins. Pull-down experiments utilize highly purified and enriched bait proteins to capture target proteins that interact weakly or are low in abundance in cells, significantly improving the efficiency of discovering new target proteins. The specific steps of the pull-down experiment are: express and purify GST, polyHis or Biotin-tagged bait proteins, use immobilized affinity ligands to stably bind the bait proteins to a solid-phase matrix, then mix the bait proteins with the test cell lysate and incubate. During this process, the bait proteins interact and bind with the target proteins. Non-specifically bound impurities are removed through washing, and the target proteins that interact with the bait proteins are then identified by mass spectrometry.
Analysis Workflow
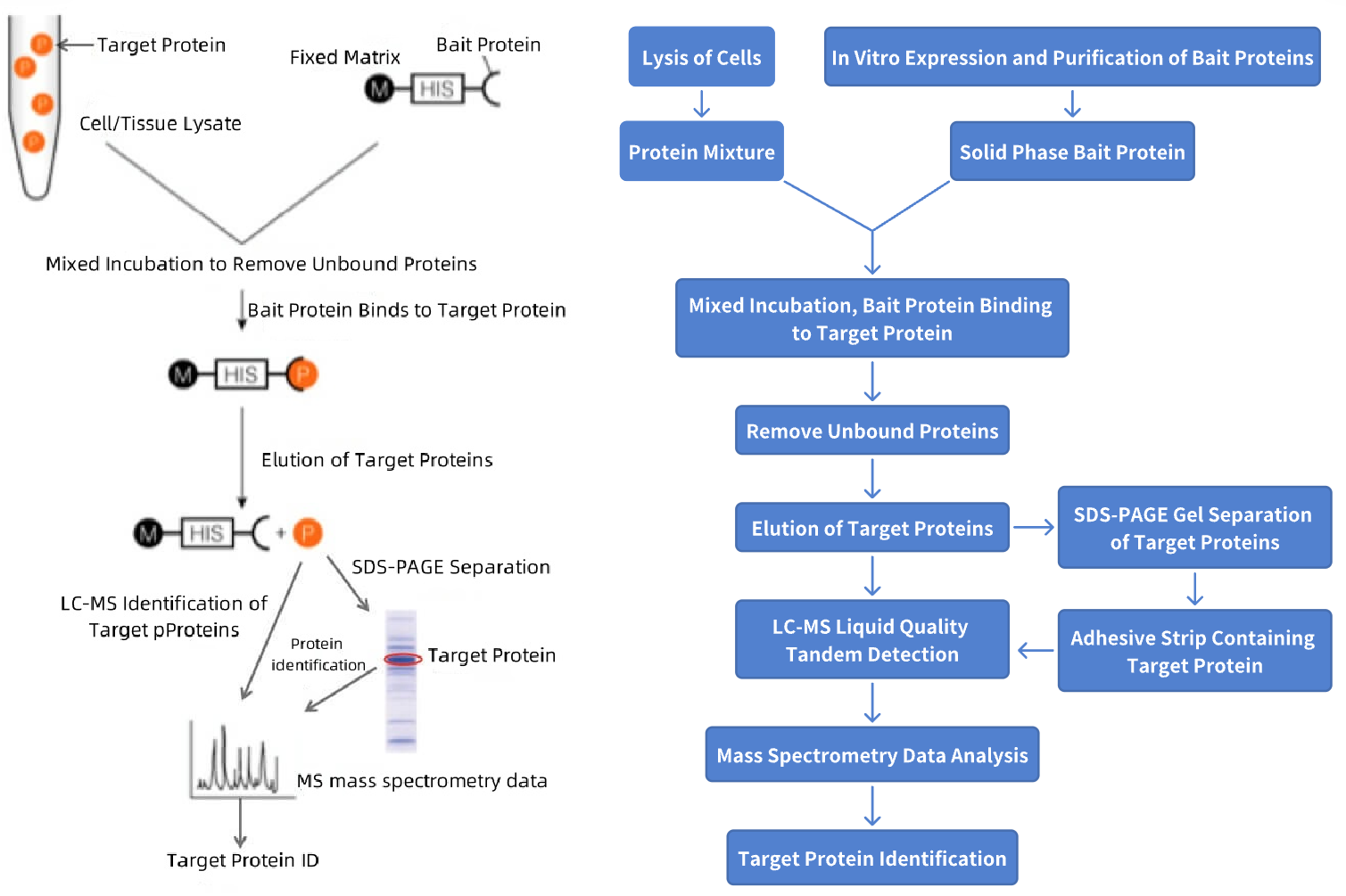
Figure 1. Pull-Down Protein Identification Process
Sample Submission Requirements
1. 1D SDS PAGE /2D PAGE Gels Samples
(1) Samples stained with coomassie blue, SYPRO Ruby, and silver stain are all suitable for protein identification experiments.
(2) Silver-stained samples may not be compatible with subsequent mass spectrometry analysis. We recommend using the following products or experimental procedures for silver staining: ProteoSilver Plus, Sigma (Product # PROTSIL1 or PROTSIL2) or Dodeca Silver Stain, BioRad (Product # 161-0481 or 161-0480).
(3) Do not decolorize silver-stained samples.
2. Liquid Samples
(1) If your eluted samples need to be run on gels, prepare them according to the requirements for 1D SDS PAGE samples.
(2) If the protein content in the sample is >10 μg, we have no requirements regarding the buffer system of the sample.
(3) If the protein content in the sample is <10 μg, try to minimize the use of surfactants such as SDS during sample preparation, and be mindful of reducing salt concentration.
(4) During the submission process, please also provide the buffer system of the sample and the estimated protein content of the sample.
3. Sample Transportation
(1) We recommend transporting samples freeze-dried, as proteins in freeze-dried samples are very stable.
(2) For liquid samples, dry ice transportation is recommended, and ice packs can also be used for short-distance transportation.
4. Precautions
When identifying protein samples using mass spectrometry, gloves and headgear should be worn during sample preparation to minimize contamination from keratin.
Deliverables
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Mass Spectrometry Images
4. Raw Data
5. Detailed Information on Identified Proteins
How to order?







