Quantitative Mass Spectrometry Service
- Labeling Quantification: Peptides in the sample are chemically labeled to distinguish them during mass spectrometry analysis. Common labeling methods include iTRAQ and TMT, which are ideal for large-scale protein quantification and allow comparison of relative abundances across multiple samples.
- Label-free Quantification: This method does not use chemical labels, instead relying on the signal intensity of peptides in mass spectrometry to calculate their relative abundance. Techniques such as SWATH and Label-free Quantification are commonly used. Label-free quantification is particularly effective for analyzing low-abundance proteins with high sensitivity, offering comprehensive analysis without the need for labeling.
- Low-complexity samples: Gel spots or bands, approximately 1-2 cm in length.
- Medium-complexity samples: Protein mixtures, including column-purified samples.
- Moderate-to-high complexity samples: Protein mixtures such as IP-derived samples, secreted proteomes, or subcellular organelles (e.g., mitochondria, endoplasmic reticulum, lysosomes).
- High-complexity samples: Whole cell lysates, tissues.
Quantitative mass spectrometry is a technique that uses mass spectrometry to precisely measure the abundance of proteins in a sample. When coupled with liquid chromatography, mass spectrometry efficiently separates the components of complex samples and provides high-sensitivity detection of target proteins, yielding detailed quantitative data. This makes it an essential tool in proteomics, clinical research, drug development, and biomarker screening. MtoZ Biolabs offers quantitative mass spectrometry service that leverage cutting-edge mass spectrometry platforms and extensive technical expertise, providing you with reliable and precise data to support both your research and commercial endeavors, ensuring outstanding results.
Technical Principles
Our quantitative mass spectrometry service enable precise measurement of protein or peptide abundance, helping clients assess changes in their relative or absolute abundance under various experimental conditions. The core methods include:
Analysis Workflow
MtoZ Biolabs’ quantitative mass spectrometry service follows a strict, standardized process to ensure each analysis meets the highest precision and reliability standards. The typical workflow consists of:
1. Sample Preparation and Digestion
After preprocessing and protein extraction, specific enzymes (such as trypsin) are used to digest proteins into peptides.
2. Labeling and Separation
For labeled quantification, peptides are chemically labeled (e.g., iTRAQ or TMT), followed by liquid chromatography (LC) separation. In label-free quantification, the labeling step is omitted, and LC separation is performed directly.
3. Mass Spectrometry Analysis
The separated peptides are introduced into the mass spectrometer, where they are ionized by electrospray ionization (ESI) and analyzed for their mass-to-charge ratio (m/z). Quantitative analysis is based on this data.
4. Data Analysis and Quantification
The mass spectrometry data is processed with specialized software to obtain quantitative results for each peptide, which are then used to calculate the relative or absolute abundance of target proteins in the sample.
Sample Submission Suggestions
For optimal outcomes in our quantitative mass spectrometry service where sample preparation and submission depend on their complexity. Proper sample handling and preparation directly influence the accuracy and reproducibility of quantitative proteomics data and enable precise protein identification and quantification across different experimental conditions. Below are our recommendations for sample submission based on complexity levels:
For more specific sample submission requirements, please consult our technical team.
Applications
Quantitative mass spectrometry service finds extensive applications across various fields, including drug target validation in drug development, biomarker screening in clinical diagnostics, and protein pollutant detection in environmental monitoring. Additionally, this technology plays a crucial role in fundamental research, enabling researchers to precisely track protein expression changes in proteomics studies, providing significant support for disease research, drug mechanism exploration, and the development of personalized medical solutions.
Case Study
Case 1
Analysis of seven transmembrane proteins involved in the DHA biosynthesis pathway in genetically engineered rapeseed using quantitative mass spectrometry.
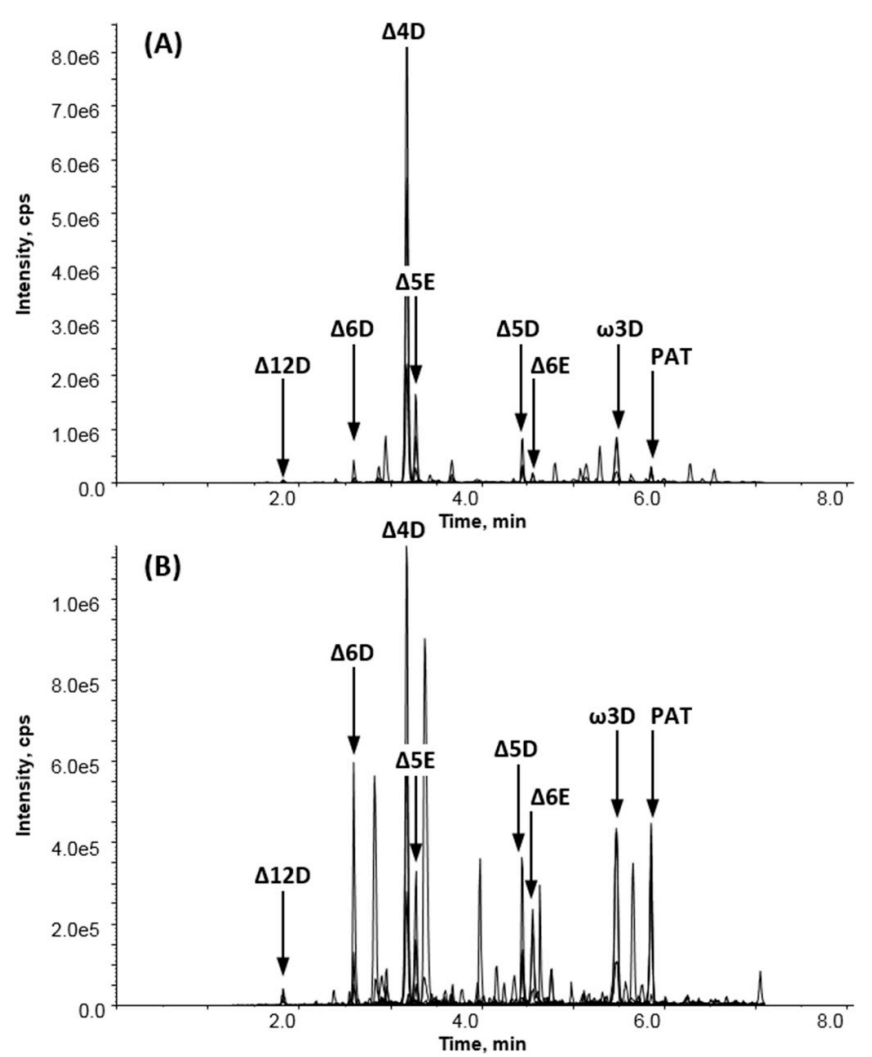
Colgrave, M. L. et al. Food Chem. Toxicol. 2019.
Case 2
Analysis of the impact of different heat treatments on the proteome of sea cucumber using label-free mass spectrometry.
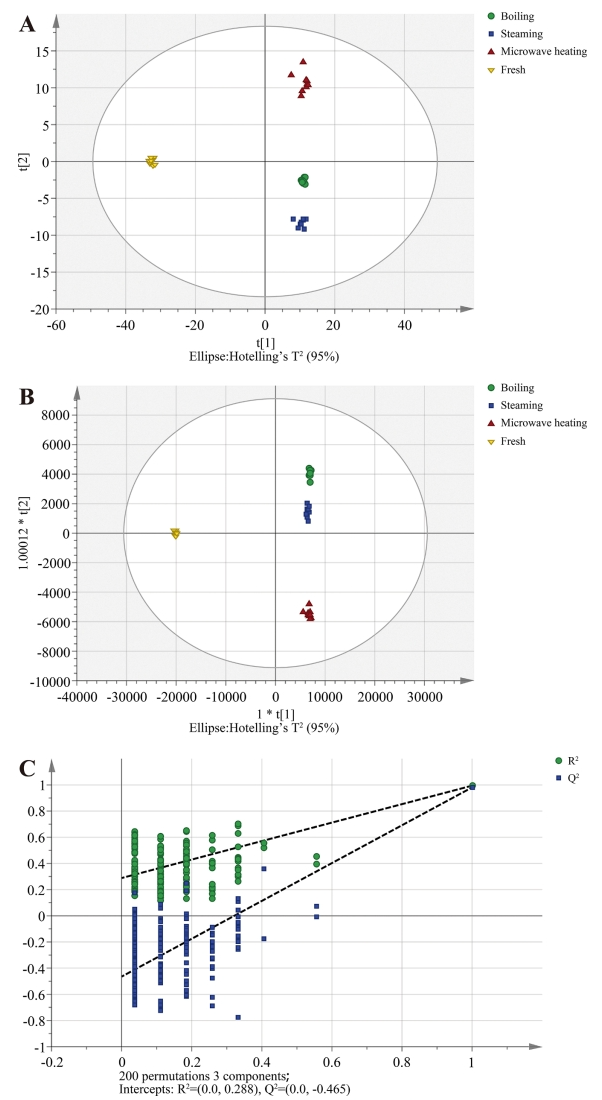
Jiang, B. X. et al. Food Chem. 2020.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our quantitative mass spectrometry service is designed to deliver rapid, high-throughput, and cost-effective analysis while ensuring exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Proteomics Service
How to order?







