Protein Sequencing Service by Mass Spectrometry
Mass spectrometry-based sequence analysis of proteins, antibodies, peptides, and other samples involves the analysis of the sample's amino acid sequence. Mass spectrometry sequencing refers to the determination of the primary structure of amino acid sequences based on mass spectrometry. Before moving on to mass spectrometry sequence analysis, we provide a brief introduction to a few definitions. Amino acids are organic compounds containing amino (-NH2) and carboxyl (-COOH) functional groups and a side chain (R group) specific to each amino acid. Peptides are short chains composed of two to fifty amino acids linked by peptide bonds. Polypeptides are longer, continuous, unbranched peptide chains, containing up to about fifty amino acids. Peptides/polypeptide chains containing more than fifty amino acids are referred to as proteins. Proteins consist of one or more polypeptides arranged in a biologically functional group manner, typically able to bind to ligands (such as coenzymes and cofactors), another protein, other large molecules (such as DNA or RNA), or more complex macromolecular assemblies. Antibodies (Ab), also known as immunoglobulins (Ig), are large Y-shaped proteins primarily produced by plasma cells, used by the immune system to neutralize pathogens, such as pathogenic bacteria and viruses.
Compared to the Edman degradation method, the advantages of mass spectrometry sequencing are that it is more sensitive, can cleave peptides faster, and can identify terminally blocked or modified protein samples. MtoZ Biolabs utilizes existing high-resolution mass spectrometry technology platforms to provide mass spectrometry-based sequence analysis services, achieving 100% coverage of the targeted protein sequence.
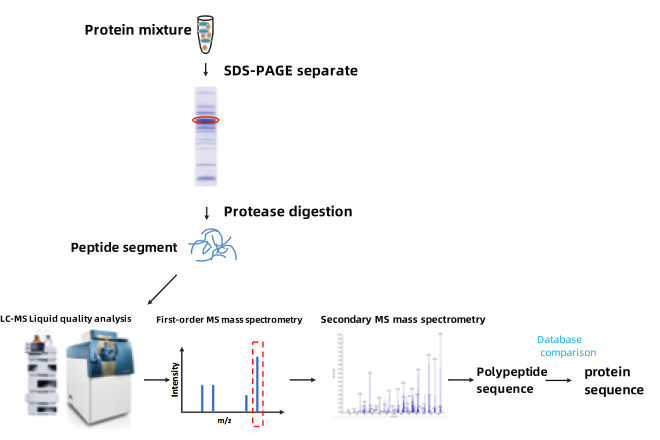
Figure 1. Mass Spectrometry Sequencing Diagram
The process of protein sample sequence analysis generally only uses Trypsin for proteolytic digestion of the protein, with peptide coverage of approximately 60%. To obtain the full sequence information of the target protein, MtoZ Biolabs selects six common proteases (Trypsin, Chymotrypsin, Asp-N, Glu-C, Lys-C, and Lys-N) used in the protein sequence analysis process to perform enzymatic digestion and mass spectrometry on the target protein. In the process of obtaining fragmented peptide fragments, the complete protein sequence is determined by splicing the peptide fragments. MtoZ Biolabs uses the Thermo company's newly launched Orbitrap Fusion Lumos mass spectrometer for protein sample sequence analysis. The Orbitrap Fusion Lumos mass spectrometer is currently the highest resolution and sensitivity mass spectrometer, ensuring the sensitivity of identification of low-abundance peptide fragmentation fragments; it also adopts a combination of HCD and ETD modes during the peptide fragmentation process to ensure the integrity of the peptide fragmentation fragments. This can achieve N-terminal, C-terminal sequence analysis, and full-length protein sequence analysis. For samples with an unknown theoretical sequence, de novo sequencing can be performed for sequence analysis.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







