Single Cell Proteomics Analysis
- Gentle sorting: Utilizing piezo-acoustic dispensing technology, which features lower shear stress and loss rate, and a higher cell survival rate.
- Optical monitoring: Advanced visual processing technology, which, through software-integrated visual feedback, automatically judges subsequent sorting operations, resulting in a high success rate.
- Low loss rate: single cell isolation rate reaches 98%, with droplets containing multiple cells being recycled for reallocation; the entire process is automated.
- Tumor heterogeneity research
- Embryonic development studies
- Immunotherapy clinical trials
- Targeted drug development
- Studies on immune mechanisms
- Resistance and dynamics studies
- Stem cell differentiation potential
- Cell atlas mapping
- Precision pathology
- Cell-specific signaling response mechanisms
- Plant single cell developmental atlases
- Stress resistance research
- single cell modifications in crop varieties
- Cell dynamics and regulatory networks
- Plant regeneration
- Comprehensive Experimental Details
- Materials, Instruments, and Methods
- Total Ion Chromatogram & Quality Control Assessment (project-dependent)
- Data Analysis, Preprocessing, and Estimation (project-dependent)
- Bioinformatics Analysis
- Raw Data Files
Single cell proteomics analysis involves examining individual cells derived from dissociated tissues for proteomics analysis, including protein composition, abundance, and modification states within single cells. Compared to conventional proteomics analysis, single cell proteomics has the advantage of providing individual cell information, enabling cell-type-specific differential expression analysis and offering revolutionary insights into cellular heterogeneity, biological system complexity, and disease mechanisms. This allows researchers to study cell behavior and function at a more refined level and is highly applicable to cancer research, immunology, developmental biology, and more.
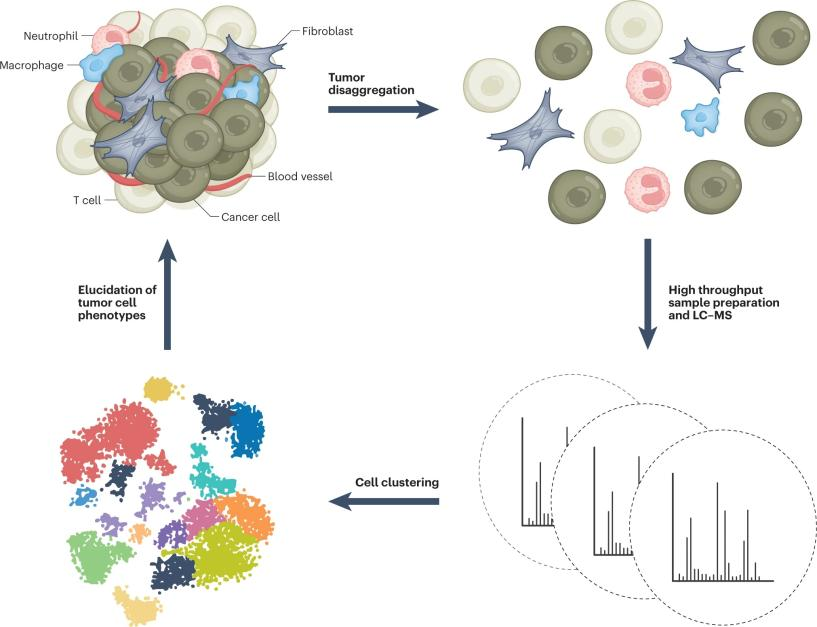
Figure1. Single Cell Proteomics Analysis (Nat Methods 20, 324–326 (2023).)
Cells are currently understood as the fundamental structural and functional units of living organisms and are the basis for all life activities. In multicellular organisms, cell variability is crucial. For example, tumors consist of various cell types, including cancer cells, fibroblasts, immune cells, and endothelial cells. Each cell type is composed of multiple different cell groups. The close interactions between these cell types often induce cell transformation within the tumor microenvironment to support tumor growth. Due to the high heterogeneity of cells, conventional proteomics cannot reflect these subtle differences. To explore the role of specific cell subpopulations in cancer development, metastasis, immune evasion, or to find genes or proteins within subpopulations as novel therapeutic targets, single cell proteomics analysis is required.
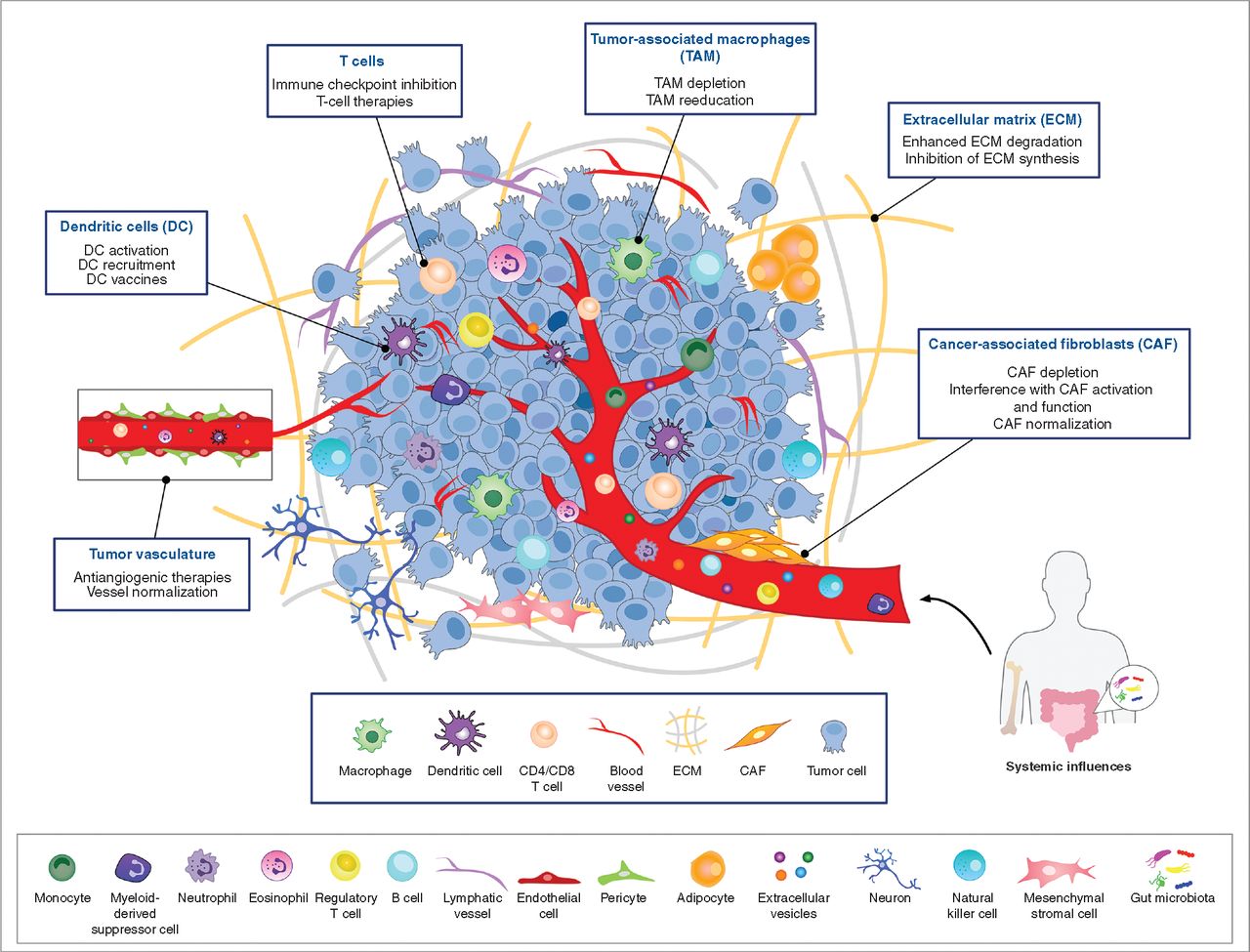
Figure2. Tumor Microenvironment (Cancer Discov (2021) 11 (4): 933-959.)
MtoZ Biolabs Single Cell Proteomics Analysis Service
MtoZ Biolabs, utilizing Thermo's advanced Orbitrap Astral high-resolution mass spectrometer and Bruker's timsTOF HT, along with the fully integrated automated single cell sorting platform CellenONE, has established a cutting-edge single cell proteomics analysis platform that achieves precise and stable detection in high-bio-sample heterogeneity scenarios:
Single cells obtained through gentle protein pre-treatment methods are enzymatically digested, and the resulting peptides are detected using the latest, highly sensitive Thermo Orbitrap Astral, ensuring data quality and providing clients with the most comprehensive single cell proteomic maps.
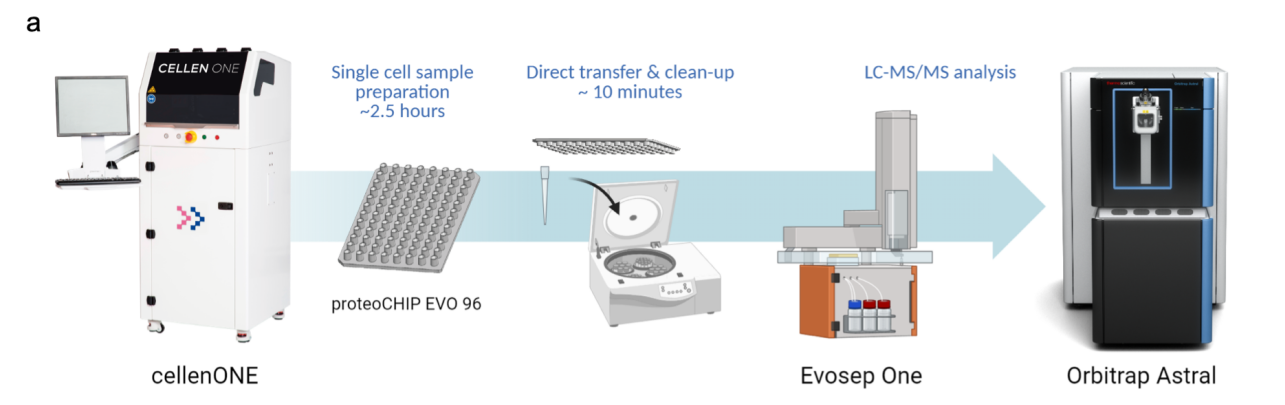
Figure3. Single Cell Proteomics Analysis Workflow
Advantages of Single Cell Proteomics Analysis
In the last decade, single cell RNA sequencing has significantly expanded our understanding of cellular heterogeneity within complex biological systems. Advances in technology now allow proteomic profiling, revealing additional cell types and states within complex tissues. Compared to single cell transcriptomics, single cell proteomics analysis offers further advantages:
1. Higher correlation with proteins
At the bulk level, transcript-protein correlation averages around 0.5, but at the single cell level, it drops to approximately 0.2 (Mol Syst Biol (2022)18: e10798).
2. Insights into protein post-translational modifications (PTMs)
PTMs increase protein diversity and regulate function, information not present in mRNA.
3. Distinct regulatory mechanisms
mRNA expression regulation and protein activity regulation differ fundamentally; transcriptomic variability often poorly predicts protein outcomes, allowing complementary insights from proteomics data.
4. Proteins are more stable, providing a closer approximation of real cell-level expression profiles
Proteomics can address heterogeneity between cells and differences between individuals, revealing cell state differences and compositional changes, uncovering physiological and pathological phenomena, and aiding in the identification of new therapeutic targets (Mol Cell. 2022;82(12):2335-2349).
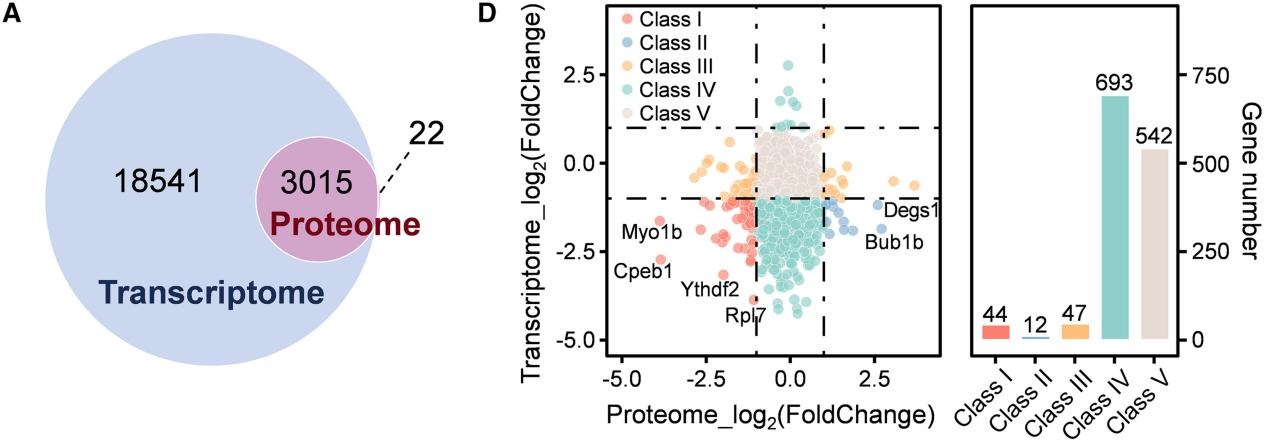
Figure4. Correlation Between Protein and Transcription (Cell Rep. 2023;42(11):113455.)
Applications of Single Cell Proteomics Analysis
In medicine:
In agriculture:
Sample Results
What Could be Included in the Report?
How to order?







