Target Identification Services
- Covalent Probes: Applicable to irreversible inhibitors, which covalently bind to the target protein through active groups (such as electrophilic groups) to improve the specificity of target recognition.
- Non-Covalent Probes: Used for reversible binding small molecules, the affinity and selectivity of the binding pocket need to be optimized to reduce nonspecific binding.
- Click Chemistry Probes: For example, ALKYNE- or AZIDE-modified probes, enabling bioorthogonal labeling through click chemistry, improving stability and capture efficiency.
- Photoaffinity Labeling (PAL) Probes: Contain photo-reactive groups (e.g., aryl diazirines) that, upon light activation, capture transient protein interactions, providing flexible target identification.
Target Identification Services refers to the process of identifying protein targets that directly interact with specific small molecules (such as drugs, chemical probes, or natural products) within complex biological systems using chemical probes and mass spectrometry. Target Identification can provide key molecular basis for understanding the mechanism of action of small molecule compounds, discovering potential drug targets, revealing signal pathways and exploring new therapeutic approaches. It is the core application of chemical proteomics in precision drug development and biological mechanism research.
Target Identification mainly relies on chemical technology combined with high-resolution mass spectrometry to capture the interaction between small molecules and proteins through chemical probes or targeted enrichment methods, and perform high-throughput identification of bound proteins. Leveraging an advanced chemical proteomics platform, MtoZ Biolabs offers Target Identification Services to help clients comprehensively analyze compound-protein interactions, providing valuable data for drug target discovery, toxicology studies, and biological function research.
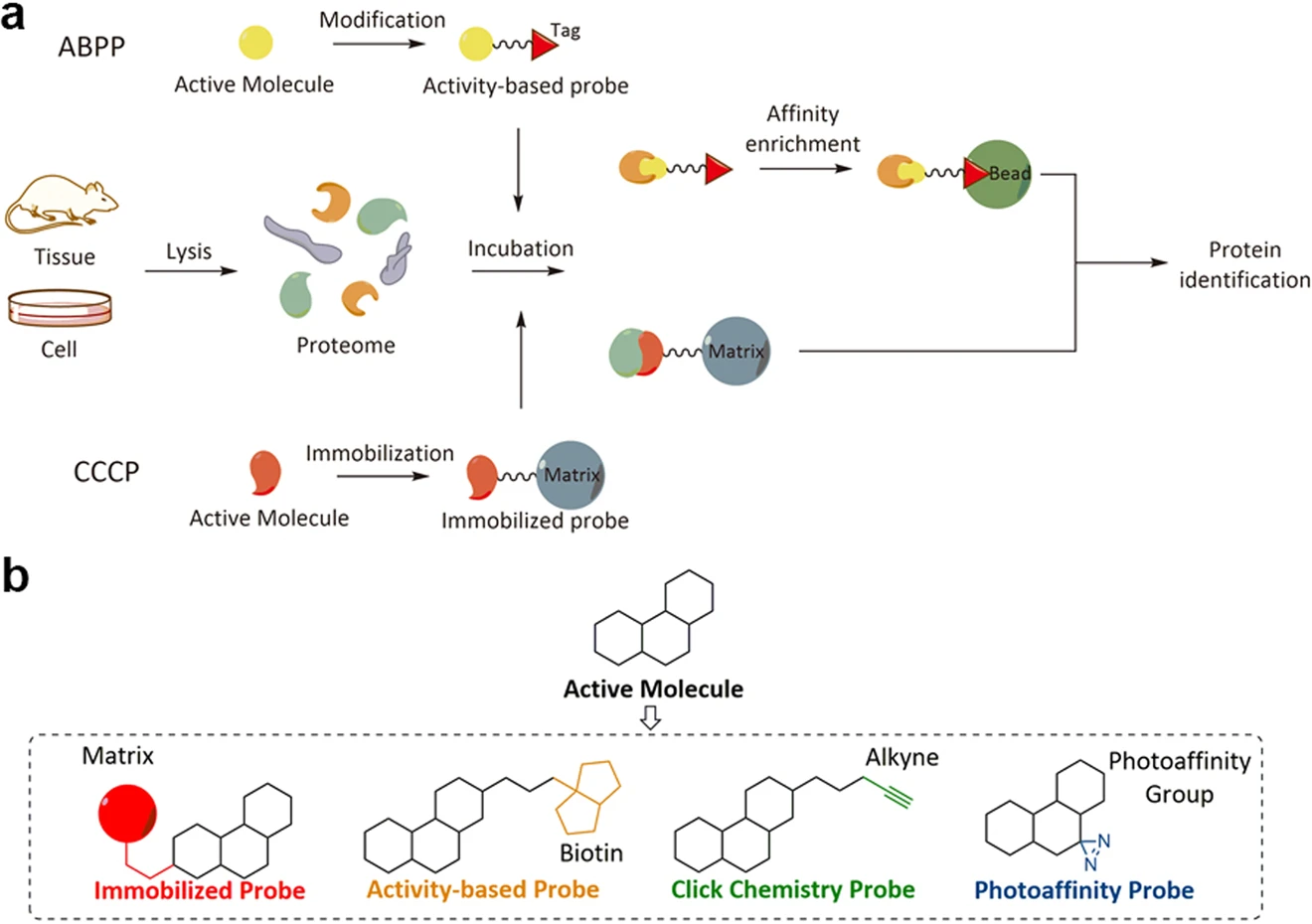
Chen X. et al. Sig Transduct Target Ther. 2020.
Figure 1. Comparison of activity-based probe profiling and compound-centric chemical proteomics
Analysis Workflow
1. Sample Preparation
Lyse cells or tissues and extract protein samples.
2. Target Labeling and Enrichment
Affinity Probe Method: Synthesize small molecule probes and incubate them with cells or protein samples to bind target proteins.
3. Mass Spectrometry Analysis (LC-MS/MS)
Perform high-resolution liquid chromatography-mass spectrometry (LC-MS/MS) for protein identification.
Precisely analyze protein sequences, post-translational modifications (PTMs), and small molecule binding sites.
4. Data Analysis and Target Validation
Match identified proteins with bioinformatics databases (e.g., UniProt, KEGG).
Utilize protein interaction network analysis tools (e.g., STRING, Cytoscape) to interpret target functions and pathways.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs provides Target Identification Services to support chemical proteomics research, covering a range of analytical approaches, including Activity-Based Protein Profiling (ABPP), MS-based thermal stability profiling, and Drug Affinity Responsive Target Stability (DARTS). For more details, please feel free to contact us.
Applications
1. Drug Target Discovery
Analyze the mechanism of action of candidate drugs and identify potential target proteins.
Detect off-target effects to optimize drug design and enhance drug safety.
2. Toxicology and Safety Assessment
Identify interactions between drugs and unintended targets to predict potential side effects.
Investigate the molecular mechanisms of environmental toxins, food additives, and chemical pollutants.
3. Metabolic Regulation and Signaling Pathway Studies
Explore how small molecules regulate protein activity and influence biological signaling pathways.
4. Biomarker Discovery and Precision Medicine
Identify disease-related protein biomarkers to facilitate precision medicine.
Analyze small molecule-protein interactions to support personalized treatment strategies.
FAQ
Q. How to Design and Select an Appropriate Chemical Probe to Efficiently and Specifically Identify Target Proteins?
The design of chemical probes should be based on the structure of the small molecule compound, target specificity, binding affinity, and feasibility of chemical modifications.
After probe design, optimization of concentration, incubation time, and elution conditions is necessary to ensure efficient binding to target proteins while reducing background noise.
Q. How to Distinguish True Target Proteins from Non-Specific Binders in Large-Scale Protein Screening?
To improve the reliability of Target Identification, multiple strategies, including experimental controls, data filtering, and bioinformatics analysis, should be employed:
1. Establish Proper Experimental Controls
Negative Controls: Use DMSO or an unmodified small molecule to eliminate non-specific binders.
Competitive Binding Assay: Introduce an excess of free small molecule during probe incubation to verify specific binding to the target protein.
Mutant Probe Control: Use an inactive probe (e.g., one lacking a reactive group) to confirm whether the modification affects binding.
2. Optimize Enrichment Strategies
Apply different elution methods (e.g., mild elution or competitive elution) to reduce non-specific interactions.
Combine MS-based thermal stability profiling or drug affinity responsive target stability (DARTS) approaches to identify target proteins based on changes in protein stability.
3. Data Analysis and Filtering
Set a False Discovery Rate (FDR) threshold (typically <1%) to eliminate low-confidence identifications.
Use label-free quantification (LFQ) to compare protein enrichment levels across samples and identify significantly enriched proteins.
Cross-validate identified proteins using protein interaction databases (e.g., STRING, BioGRID) to confirm their relevance to known targets.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized Target Identification to identify proteins in Herpes Simplex Virus Type 1 (HSV-1) associated with neurovirulence. The results demonstrated that the ICP34.5 protein in HSV-1 plays a crucial role in host cells by interacting with multiple host proteins, thereby influencing the virus’s pathogenicity and neurotoxicity. Through chemical proteomics, the study screened and identified key viral virulence factors, providing critical insights into HSV-1 pathogenesis and potential antiviral target development.
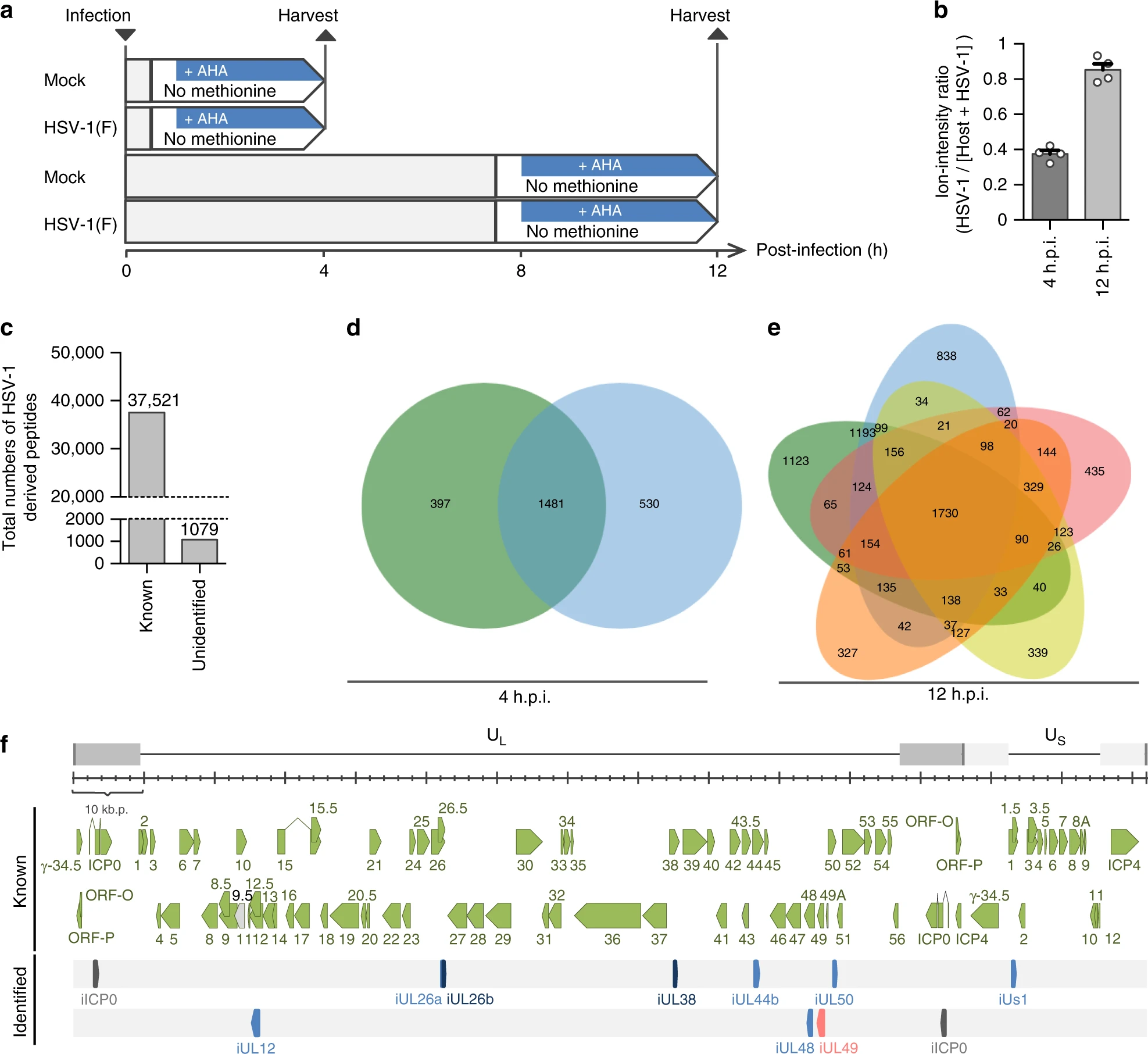
Kato A. et al. Nature Communications. 2020.
Figure 2. Identification of previously unidentified HSV-1 CDSs by chemical proteomics
How to order?







