Kinome Profiling Service
Kinome profiling is a cutting-edge analytical approach that provides a comprehensive overview of kinase function, activity, expression, and regulatory networks within a biological system. The kinome, representing the complete set of protein kinases encoded by the genome, is central to cellular signaling and the regulation of numerous biological processes. Protein kinases mediate signal transduction by catalyzing the phosphorylation of target substrates, a reversible modification that influences protein activity, localization, and interactions. Dysregulation of kinase activity is implicated in various diseases, including cancer, autoimmune disorders, and neurodegenerative conditions. Understanding the kinome is essential for deciphering the molecular mechanisms behind these conditions and addressing the underlying biological complexity.
Kinome profiling plays a pivotal role in drug discovery and target validation, elucidates disease mechanisms at the molecular level, and facilitates biomarker identification for diagnostic and therapeutic purposes. Kinome profiling has become indispensable for both basic research and translational studies, advancing personalized medicine and precision therapeutics.
Service at MtoZ Biolabs
Recognizing the critical need for high-quality kinome analysis, MtoZ Biolabs offers a comprehensive Kinome Profiling Service. Our Kinome Profiling Service integrates advanced mass spectrometry, phosphorylation-specific enrichment, and bioinformatics to quantitatively map kinase activity and detect post-translational modifications. By enabling high-throughput analysis of active kinases, kinome profiling helps unravel complex signaling cascades, identify key nodes of regulation, and reveal potential drug targets.
Our Kinome Profiling Service provides researchers with highly accurate kinase activity quantification, substrate identification, and detailed pathway insights. Whether for pharmaceutical development, pathway elucidation, or biomarker discovery, our Kinome Profiling Service is tailored to deliver actionable data and empower your research endeavors.
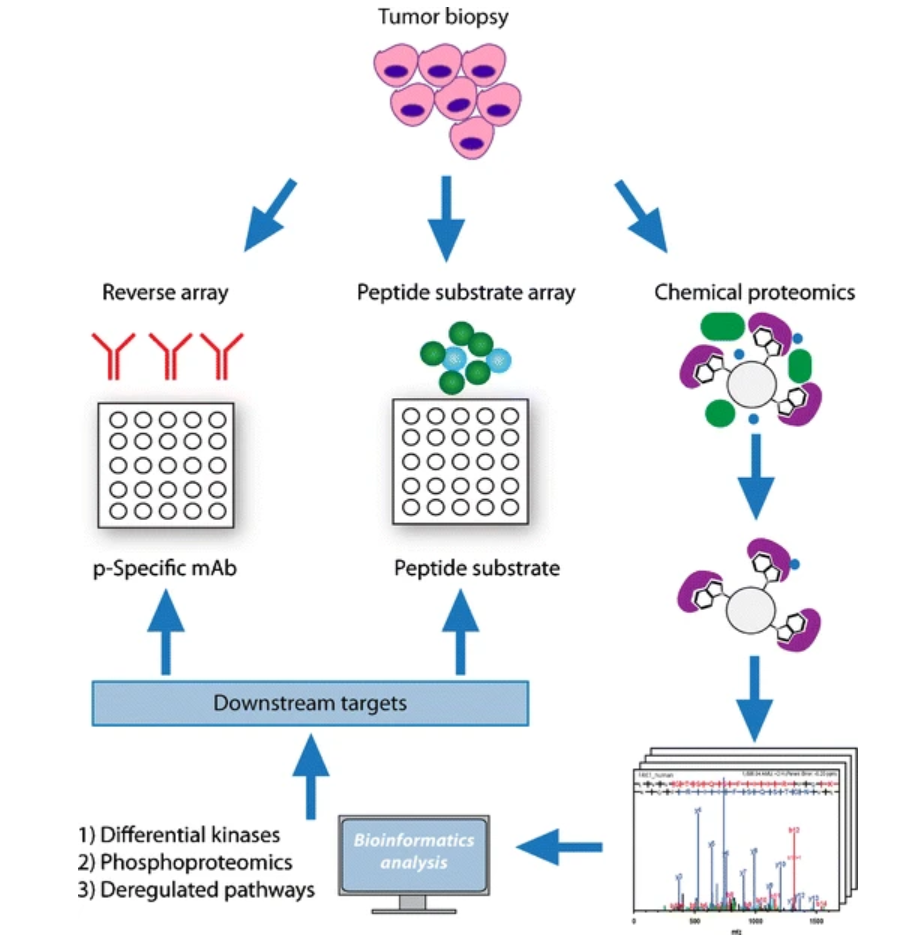
Figure 1. Overview of Kinome Profiling Approaches
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Kinome Profiling Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. Comprehensive Kinase Analysis: The combination of chemical proteomics and high-resolution LC-MS/MS allows for the identification of kinase abundances, phosphorylation sites, and interacting proteins, providing a holistic view of kinase signaling networks.
3. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
4. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Kinome Profiling Service data, providing clients with a comprehensive data report.
5. Versatile and Flexible Applications: Suitable for various sample types, including cell lysates, tissue extracts, or clinical samples, the workflow can be customized to address specific research needs in drug discovery, target validation, and disease mechanism studies.
Case Study
1. Kinome Profiling of Primary Endometrial Tumors
Endometrial carcinoma (EC), the most common gynecologic malignancy, exhibits frequent kinase alterations, yet few have been explored as therapeutic targets. To uncover potential therapeutic targets, kinome profiling was performed on endometrial tumors (n=20) and normal tissues (n=16) using Multiplexed Inhibitor Beads coupled with Mass Spectrometry (MIB-MS). Figure A illustrates the workflow, where kinases were enriched and quantified using Label-Free Quantification (LFQ) and validated with super-SILAC spiked-in standards. The study identified 347 kinases, with significant overexpression observed in tumor tissues compared to normal controls (Figure B). On average, LFQ detected a higher number of kinases per sample, achieving robust coverage across >90% of MIB-MS runs (Figures C, D). Hierarchical clustering revealed distinct kinase expression patterns between EC and normal tissues, highlighting dysregulated kinases enriched in tumors (Figure E). Principal Component Analysis (PCA) further confirmed the segregation of tumor samples from normal tissues, emphasizing kinome differences (Figure F). Key findings identified SRPK1 as an overexpressed kinase, correlating with poor survival in EC patients. Targeting SRPK1, combined with EGFR/IGF1R inhibition, synergistically suppressed cell growth, demonstrating the therapeutic potential of dual kinase targeting. This case study showcases the power of Kinome Profiling in uncovering actionable kinases, guiding the development of combination therapies to improve outcomes in EC.
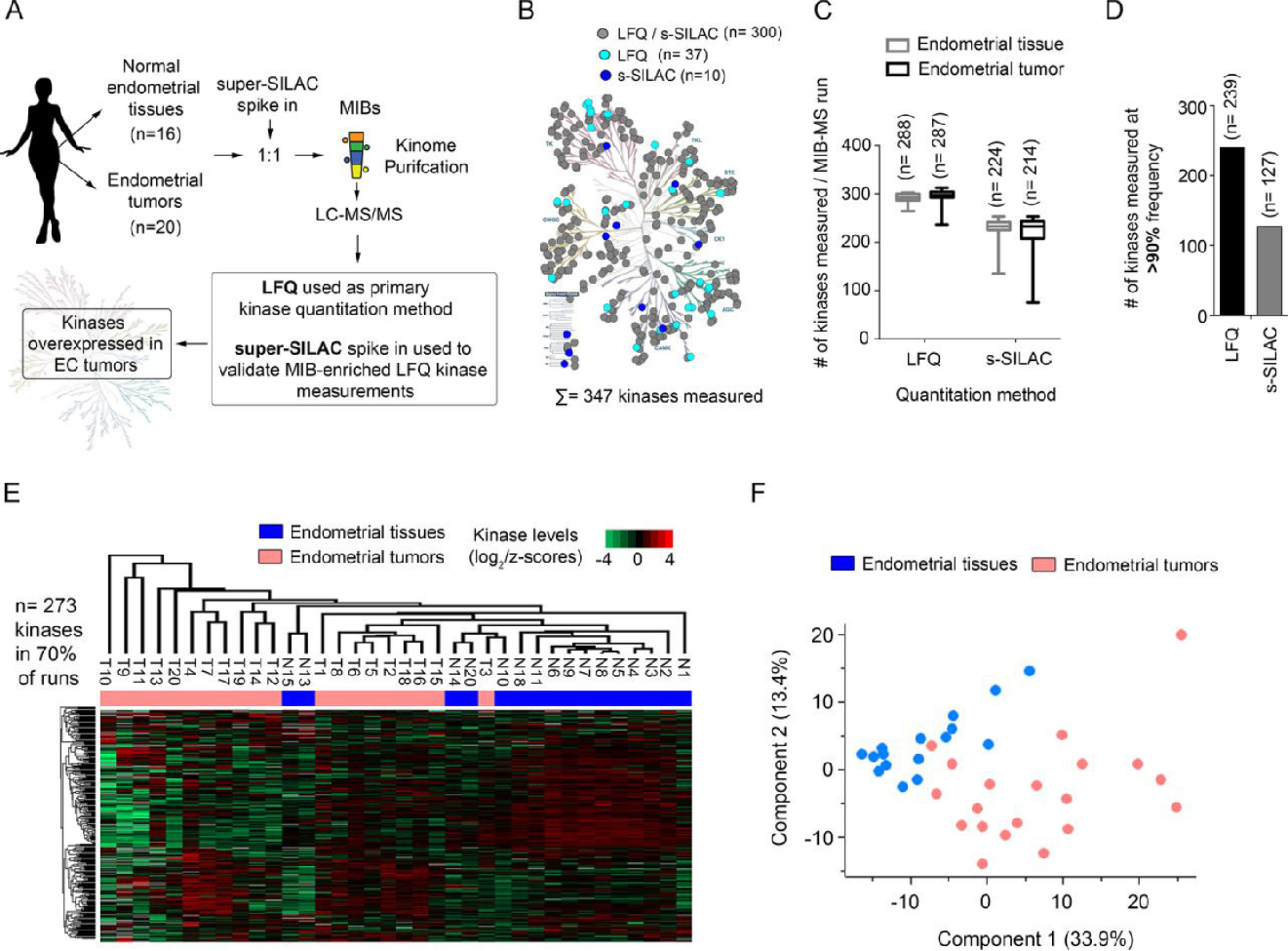
Figure 2. Kinome Profiling of the Endometrial Cancer
2. Subcellular Active Kinome Profiling
Protein kinases regulate signaling networks within specific subcellular compartments, yet comprehensive activity profiling at the synaptic level remains limited. In this study, Kinome Profiling was performed on rat frontal cortex tissue to analyze kinase activity across nuclear, cytosolic, and synaptosomal fractions. Subcellular fractionation and enrichment were validated using Western blotting, confirming the purity of each fraction. Using peptide arrays on the PamStation12 platform, global serine/threonine kinase activity was assessed. The results revealed distinct kinase activity profiles across compartments, with hierarchical clustering identifying differentially active kinases enriched in the synaptosomal fraction. Novel kinases with previously unknown synaptic localization were also discovered, alongside those already established in synaptic signaling. This study highlights the value of Kinome Profiling in identifying compartment-specific kinase activity, advancing our understanding of signaling networks in distinct cellular fractions and uncovering new potential targets for research and therapy.
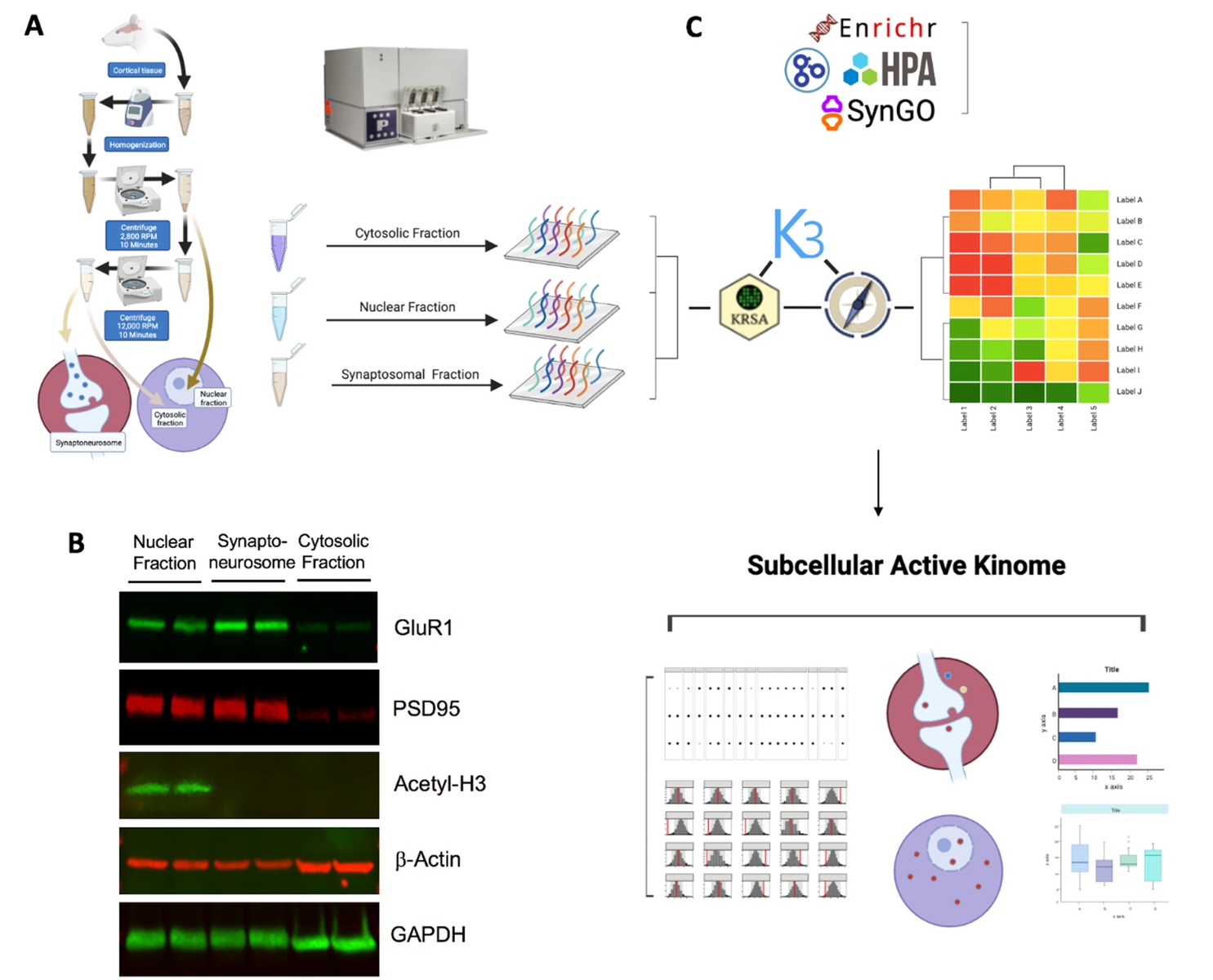
Wegman-Points, L. et al. Sci Rep. 2022.
Figure 3. Subcellular Active Kinome Profiling
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
How to order?







