Native Protein Analysis Service
- Molecular weight determination of proteins and protein complexes in their native state.
- Structural and topological analysis of native proteins and protein complexes.
- DAR assays for antibody-drug conjugates (ADCs) analysis.
- Analysis of PEG or polysaccharide-modified proteins for bioconjugate characterization.
- Protein or protein–drug complex assembly studies.
Accurate molecular weight determination is a cornerstone of protein characterization, providing essential information about protein size, oligomeric state, and structural integrity. The Native Protein Analysis Service offered by MtoZ Biolabs focuses specifically on determining the molecular weight of proteins in their native, non-denatured state, ensuring that the data reflects the protein's true conformation and functional integrity.
Understanding proteins in their native state is crucial for gaining accurate insights into their structure, function, and interactions. Native protein analysis allows for the study of intact proteins and protein complexes—such as monoclonal antibodies—while preserving their true biological conformation. By analyzing proteins without denaturation, we can generate a comprehensive "fingerprint" of the sample, revealing subtle differences between proteins, including post-translational modifications (PTMs) and variations in protein isoforms.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
At MtoZ Biolabs, our Native Protein Analysis Service focuses on precise molecular weight determination of proteins in their natural state using advanced techniques like liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS). This approach reduces sample complexity and preparation time compared to peptide-based proteomics methods, enabling more efficient and accurate profiling of proteins in complex samples. Our sophisticated data processing algorithms utilize retention time and exact mass measurements to identify and characterize proteins, facilitating statistical analysis and visualization for differential protein studies. Whether you're investigating protein structure, conformational changes, or PTMs, our native protein analysis provides high-quality, reproducible data essential for advancing your research and development objectives. If you are interested in our service, please feel free to contact us.
Analysis Workflow
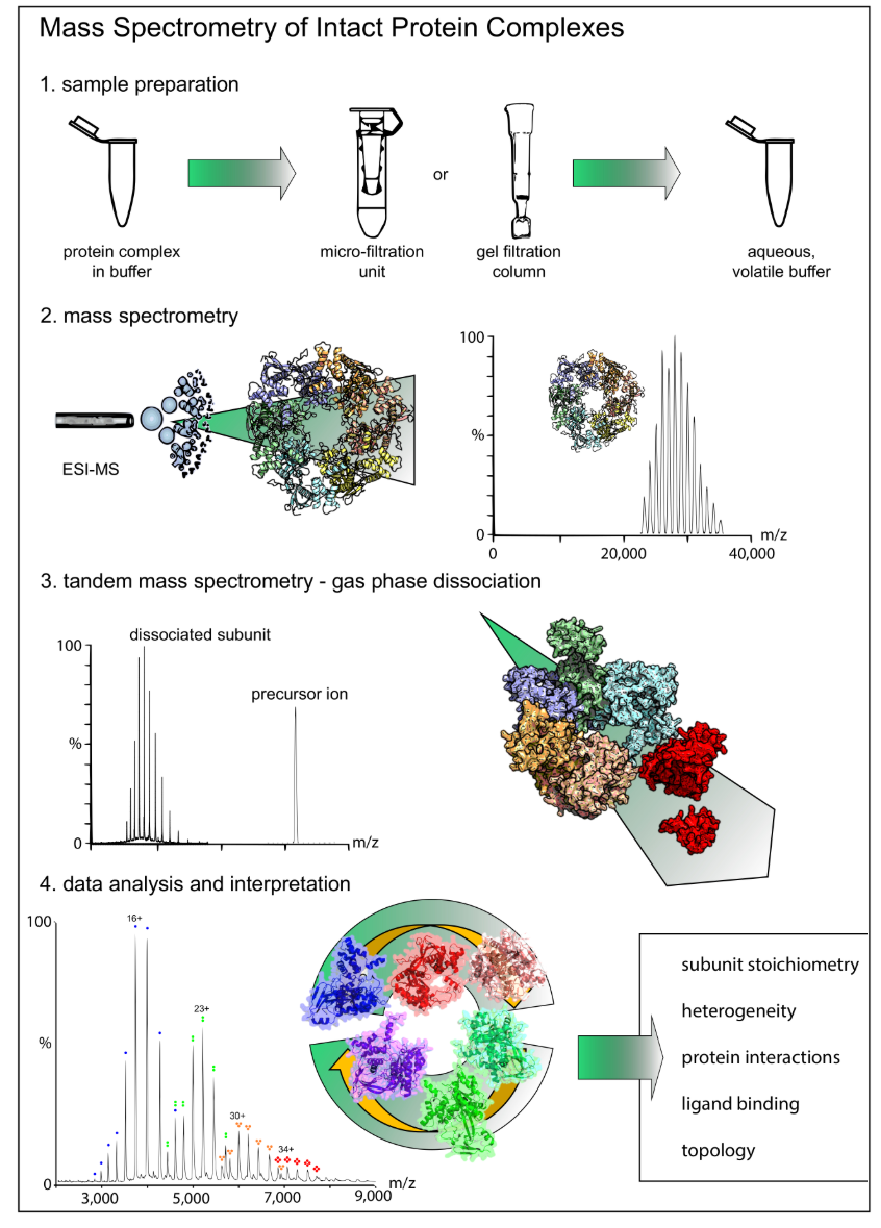
Figure 1. Workflow for Native Protein Analysis Service
Why Choose MtoZ Biolabs?
1. Advance Analysis Platform
MtoZ Biolabs established an advanced Native Protein Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality
Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all native protein analysis data providing clients with a comprehensive data report.
4. Simplified Sample Preparation
This approach reduces sample complexity and minimizes preparation time, ensuring efficient and accurate profiling of proteins in complex samples.
5. Accurate Native Profiling
By analyzing proteins in their native state, we preserve their structural integrity, allowing for precise detection of post-translational modifications (PTMs) and protein isoforms.
Applications
Case Study
As shown in the figure below, we observed higher-order oligomers up to 16-mers at m/z of ∼18,000, corresponding to a molecular weight of 1.86 MDa. Macromolecular protein complex with molecular weights up to 1.86 MDa can be transmitted and detected by FTICR mass spectrometry under native ESI condition.
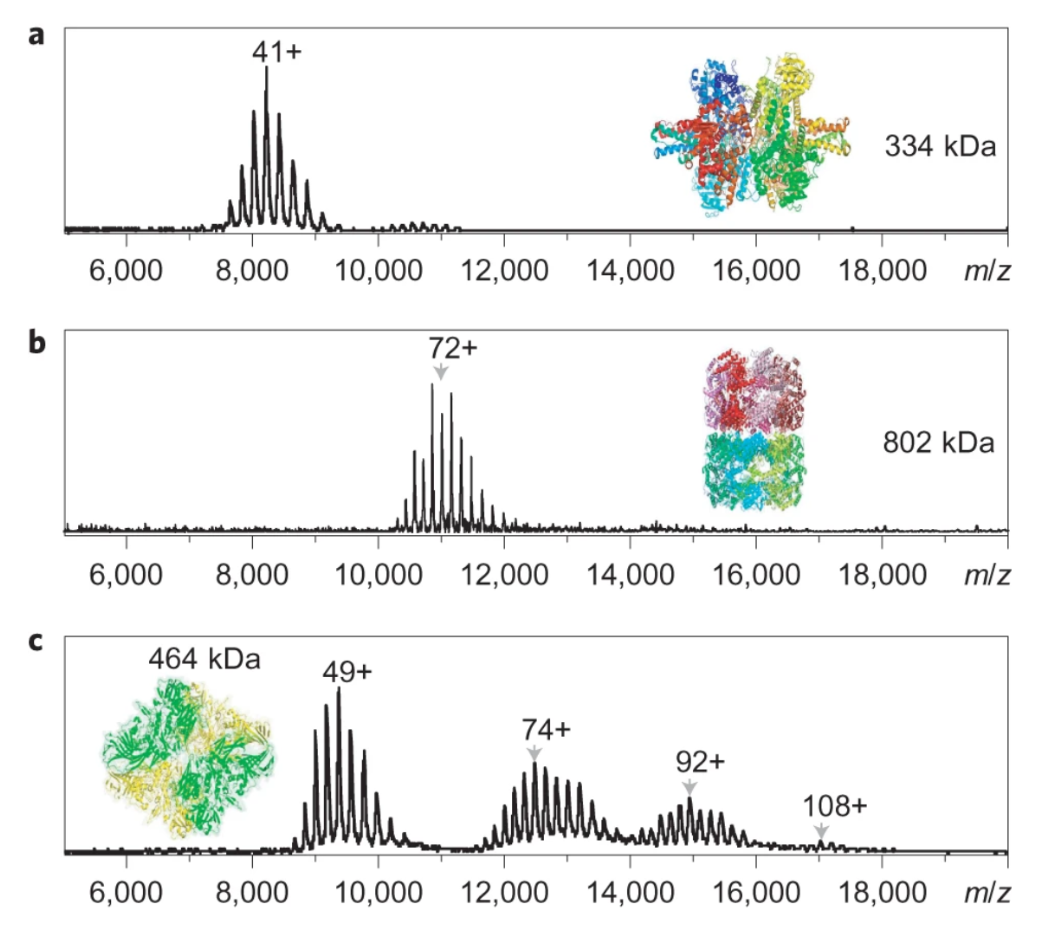
Li, H. et al. Nat Chem. 2018.
Figure 2. Macromolecular Protein Complexes Transmitted and Detected by FTICR Mass Spectrometry Under Native ESI Condition
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Mass Spectrometry Parameters
4. Detailed Information on Native Protein Molecular Weight
5. Mass Spectrometry Images
6. Raw Data Files
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Native N-glycome profiling Service
How to order?







