By combining MALDI-TOF mass spectrometry system and LC-MS/MS mass spectrometry system, MtoZ Biolabs provides professional and accurate protein identification services for protein spots, strips, and IP samples, as well as Co-IP sample proteins. The identifiable samples include protein strips separated by 1D-SDS PAGE, protein spots in 2DE PAGE, protein expression products, and Co-IP samples. The pre-identification process of protein samples strictly follows the article published in Nature Methods: "Universal sample preparation method for proteome analysis," ensuring a higher peptide coverage rate in the identification experiment. The principle of mass spectrometry identification is based on the shotgun approach: protein bands or protein solutions are enzymatically digested into a mixture of peptides, which are then separated by HPLC to obtain several relatively simple components for analysis by high-resolution mass spectrometry. Retrieval software compares the secondary mass spectrometry information with the corresponding database, combines matching scores and mismatch filtering, and obtains the exact sequence of peptides, thereby splicing the complete sequence of each protein and achieving protein identification.
Sample Submission Requirements
1. 1D SDS PAGE / 2D PAGE Gels Samples
(1) Coomassie blue, SYPRO Ruby, and silver stain samples can all be subjected to protein identification experiments.
(2) Silver-stained samples may be incompatible with subsequent mass dpectrometry analysis. We recommend the following products or experimental procedures for silver staining experiments.
① ProteoSilver Plus, Sigma (Product # PROTSIL1 or PROTSIL2)
② Dodeca Silver Stain, BioRad (Product # 161-0481 or 161-0480)
(3)Do not bleach silver stain samples.
2. Liquid Samples (In Solution Format)
(1) If the protein amount in the sample is >10 μg, there are no specific requirements for the buffer system of the sample.
(2) If the protein amount in the sample is <10 μg, try to minimize the use of SDS and other surfactants in the sample preparation process, and pay attention to reducing the salt concentration.
(3) During sample submission, please provide the buffer system of the sample and the estimated protein content in the sample.
3. Co-IP Samples
(1) If the eluted samples need to be run on gels, prepare the samples according to the requirements for 1D SDS PAGE samples.
(2) If you directly submit eluted samples, we recommend using the HPH EB (0.5 M NH4OH, 0.5 mM EDTA) buffer system for protein elution to ensure compatibility with subsequent mass spectrometry experiments.
4. Sample Transportation
(1) We recommend transporting freeze-dried samples, as proteins in freeze-dried samples are very stable.
(2) For liquid samples, dry ice transportation is recommended, and ice packs can also be used for short-distance transportation.
5. Precautions
When using mass spectrometry for protein sample identification, wear gloves and a head covering during sample preparation to minimize contamination from keratin.
Case Study
This project involves a teacher from the Institute of Biophysics, who sent her Co-IP samples to us for protein identification. Some of the results obtained are shown in the figure below:
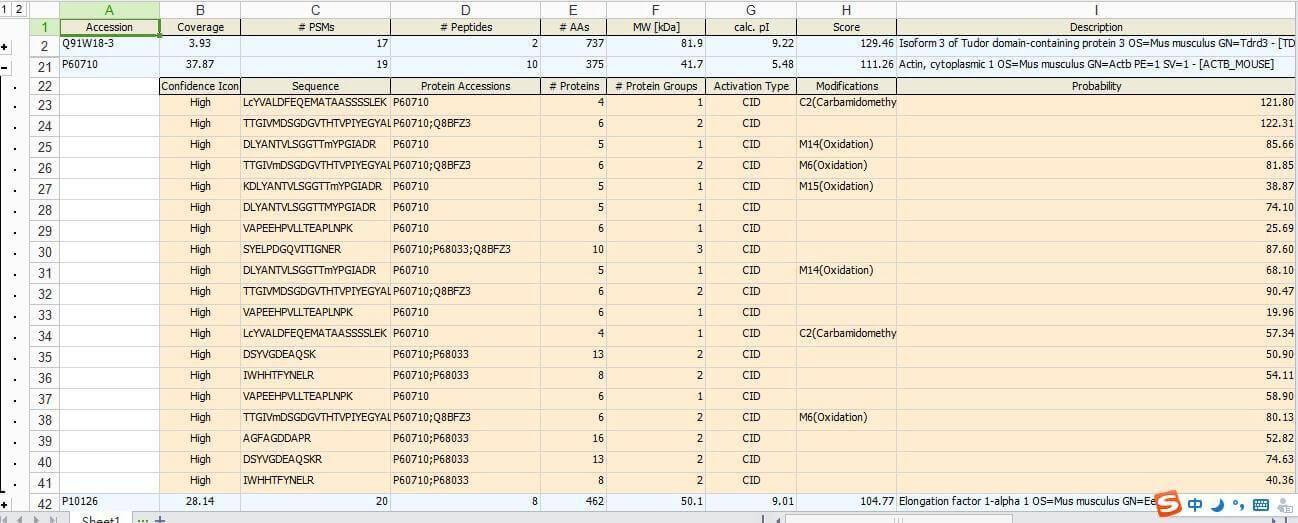
Figure 1. Case Study of Protein Spot, Strip, and IP Sample Protein Identification
From the figure, it can be seen that the results report we provide includes the accession number, peptide coverage, identified peptide information, modified information of identified peptides, and a detailed description of the identified proteins.
Deliverables
In the technical report, MtoZ Biolabs will provide you a detailed information, including:
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Mass Spectrometry Images
4. Raw Data
5. Detailed Information on Identified Proteins








