Protein Sequencing Service
- Protein Purity Assessment: Verifies whether the sample's purity meets experimental requirements through amino acid composition data.
- Expression and Processing Validation: Evaluates whether recombinant protein expression resulted in unexpected modifications or degradation.
- Functional Domain Prediction: Infers potential functional domains based on the proportion of specific amino acids.
- Edman Degradation: Ideal for high-purity samples, this method sequentially releases and identifies amino acid residues, ensuring direct sequence resolution.
- Mass Spectrometry-Assisted Approach: Employs LC-MS/MS technology in conjunction with specific enzymatic digestion strategies and targeted enrichment of N-terminal modifications for rapid analysis of complex samples.
- Specific Enzymatic Digestion: Utilizes carboxypeptidases or specific enzymatic cleavage combined with mass spectrometry to precisely resolve C-terminal sequences.
- C-terminal Modification Detection: Identifies post-translational modifications like carboxyl methylation or esterification and explores their roles in regulatory functions.
- Complex Sample Analysis: Employs high-resolution mass spectrometry and bioinformatics tools to eliminate interference signals and ensure accurate C-terminal sequence resolution in mixed protein samples.
- Multi-Enzyme Digestion Strategies: Design customized digestion schemes using enzymes such as trypsin, Lys-C, and Glu-C to generate fragments that cover the entire protein sequence.
- LC-MS/MS Analysis: Use advanced mass spectrometry instruments (e.g., Orbitrap Fusion Lumos) to perform precise analyses of peptide fragment masses and secondary fragmentation data.
- Sequence Assembly and Validation: Employ proprietary algorithms to reconstruct the complete sequence from peptide fragments and validate it against reference databases such as UniProt.
- Comprehensive PTM Analysis: Simultaneously identify modifications such as phosphorylation, glycosylation, and methylation, creating a complete functional map of the protein.
- Advanced Mass Spectrometry Configuration: Uses high-resolution instruments such as FTICR-MS to ensure data accuracy and dynamic range.
- Optimized Digestion and Derivatization Strategies: Generates peptide fragments with controlled digestion and enhances signal intensity through chemical derivatization.
- Bioinformatics Tools Support: Applies proprietary algorithms for secondary mass spectrometry data interpretation, leveraging probabilistic models to deduce amino acid arrangements and reconstruct complete sequences.
- Unknown PTM Analysis: Identifies novel modification sites and investigates their potential roles in regulating protein activity.
Protein sequencing refers to the high-precision determination of the amino acid sequence of protein molecules and is a core technology in molecular biology, structural biology and proteomics. By deciphering the primary structure of proteins, researchers can uncover their functions, regulatory mechanisms, and roles within complex biological systems. The protein sequencing Service is essential for providing the detailed sequence data needed for a comprehensive understanding of protein function in a variety of biological contexts.
Figure1. Protein Amino Acid Sequence
The origins of protein sequencing can be traced back to the 1950s, when Fred Sanger developed the insulin sequencing method, laying the foundation for modern protein science. However, traditional techniques, such as Edman degradation, have limitations, including high sample requirements, operational complexity, and restricted resolution, making them inadequate for contemporary scientific research. In contrast, the advanced protein sequencing Service offered by MtoZ Biolabs utilizes state-of-the-art methods to overcome these challenges, providing high-accuracy results for complex protein analyses.
With the advent of mass spectrometry, liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based protein sequencing has emerged as the dominant approach. This technique employs multi-enzyme digestion strategies to generate peptides, measures their masses using mass spectrometry, and determines the complete amino acid sequence by database matching and algorithmic interpretation. It not only enables the analysis of low-abundance proteins but also facilitates the simultaneous identification of post-translational modifications (PTMs), significantly expanding the scope of protein research. The protein sequencing Service at MtoZ Biolabs makes use of this powerful technology to deliver detailed insights into protein structure and function. It not only enables the analysis of low-abundance proteins but also facilitates the simultaneous identification of post-translational modifications (PTMs), significantly expanding the scope of protein research.
Services at MtoZ Biolabs
As a global leader in mass spectrometry analysis services, MtoZ Biolabs provides protein sequencing service based on Nano Liquid Chromatography-Tandem Mass Spectrometry (Nano LC-MS/MS) and Shimadzu’s Edman degradation system, which can achieve amino acid composition analysis, N-terminal sequencing, C-terminal sequencing, de novo sequencing, and full-length sequence analysis. For proteins with unknown sequences, de novo sequencing is available. Whether for identifying novel proteins or analyzing complex protein interactions, MtoZ Biolabs' protein sequencing Service ensures that researchers gain the most accurate and reliable data for their studies.
1. Amino Acid Composition Analysis
Amino acid composition analysis serves as a foundational step in protein research, aiming to quantitatively determine the types and proportions of amino acids present in a sample. This analysis is particularly useful in the following scenarios:
We employ acid hydrolysis to break down proteins into individual amino acids, followed by precise quantification using high-performance liquid chromatography (HPLC) and mass spectrometry. Additionally, specific modifications such as hydroxyproline and methionine oxidation can be identified, providing supplementary insights for post-translational modification (PTM) studies.
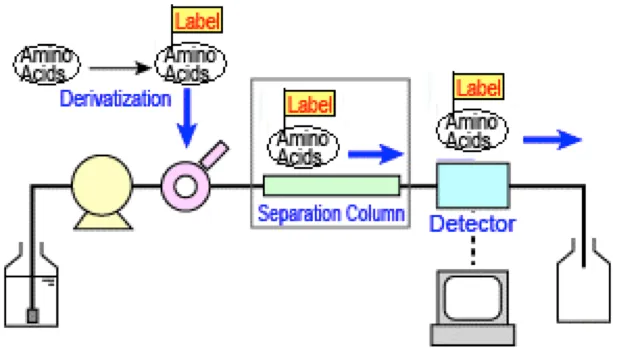
Figure2. Amino Acid Composition Analysis
2. N-terminal Sequencing
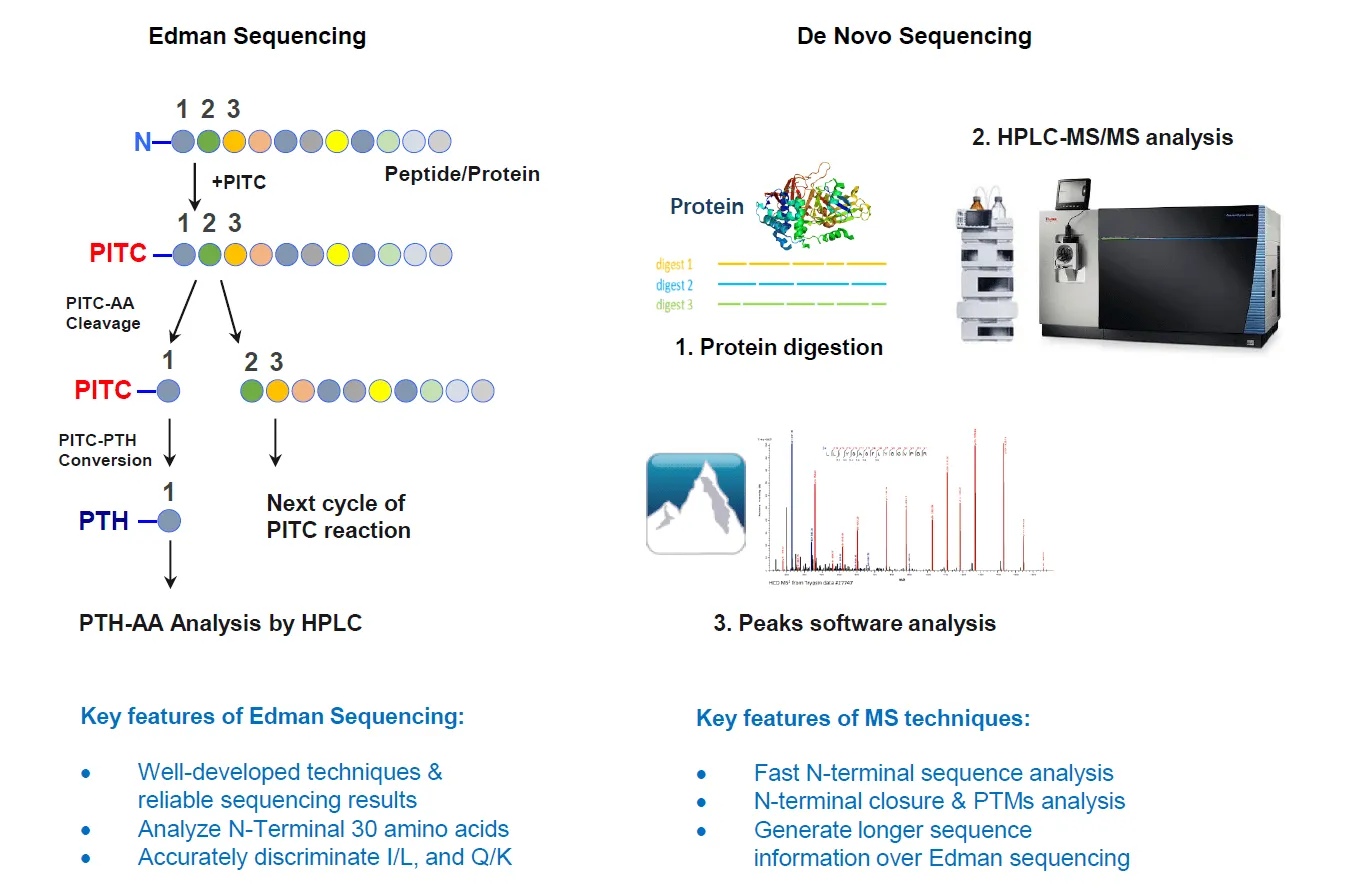
N-terminal sequencing is the starting point for determining protein amino acid sequences and holds particular significance in antibody development and protein function studies. While traditional methods like Edman degradation offer high precision, they pose challenges in terms of sample quantity requirements and operational complexity. At MtoZ Biolabs, we perform N-terminal sequencing using the following methods:
This protein sequencing service allows us to obtain high-quality N-terminal sequence data for both purified proteins and complex samples, enabling us to identify protein isoforms, confirm protein expression, and assess modifications at the N-terminus. By combining both traditional and mass spectrometry-assisted techniques, we are able to provide comprehensive and reliable sequencing results that meet the needs of diverse proteomics projects.
Figure3. Protein N-Terminal Sequencing Workflow Based on Mass Spectrometry and Edman
3. C-terminal Sequencing
C-terminal sequencing is another critical service offered in the protein sequencing service suite, allowing for the precise determination of the C-terminal amino acid in proteins. C-terminal sequence information is crucial for understanding the complete structure of a protein. However, challenges such as low enzymatic efficiency and complex modifications often complicate its detection. MtoZ Biolabs has developed efficient C-terminal sequencing strategies to accurately identify terminal amino acids and related modifications:
Through C-terminal sequencing, we provide clients with complete protein structural information, facilitating in-depth studies of protein function, activity, and processing mechanisms.
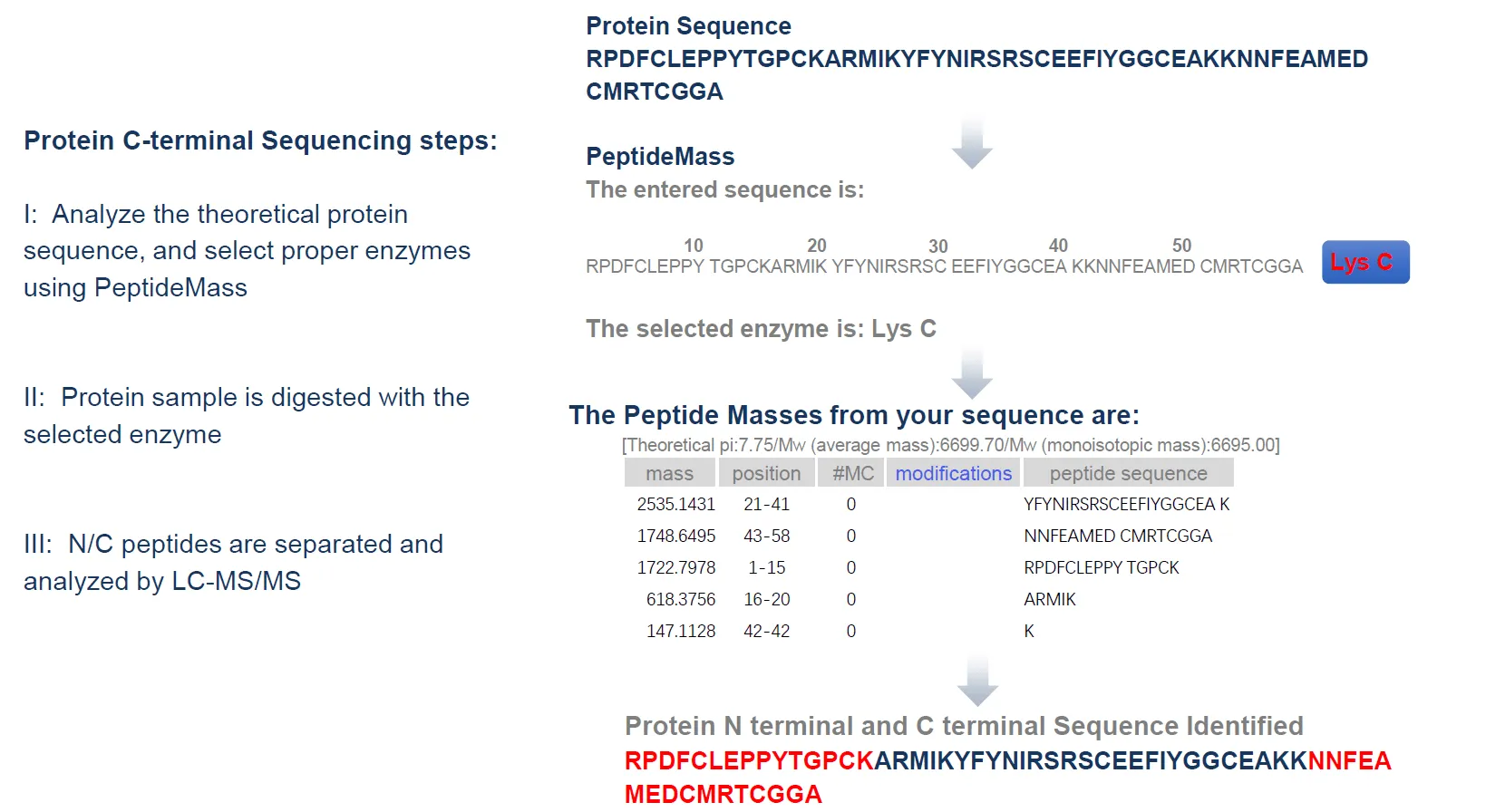
Figure4. Protein C-Terminal Sequencing Workflow
4. Full-Length Protein Sequencing
Full-length protein sequencing is a comprehensive approach to resolving the complete structure of proteins. This technique is widely used in novel protein discovery, antibody drug development, and protein engineering. MtoZ Biolabs' protein sequencing service utilizes high-resolution mass spectrometry and a multi-enzyme digestion strategy to provide high coverage, highly accurate, full-length sequence information. Key steps include:
Our full-length sequencing services are extensively applied in antibody engineering optimization, industrial enzyme modification, and novel protein functionality studies, driving breakthroughs in life science research.
5. De novo Sequencing
De novo sequencing is a powerful tool within our protein sequencing Service, especially useful when working with proteins whose sequences are unknown or unannotated in protein databases. Using cutting-edge LC-MS/MS technology, we analyze peptide fragments generated by enzymatic digestion to determine the protein’s complete amino acid sequence. This method provides unparalleled flexibility for identifying novel proteins or studying organisms without complete genome sequences.
De novo sequencing provides robust support for functional studies of unknown proteins, comparative analysis across species, and evolutionary biology research. It has also become a crucial tool for discovering biomarkers.
Sample Submission Requirements
1. Sample Preparation
There are multiple methods for sample preparation, each with its own advantages and disadvantages. The optimal method can be determined based on laboratory experience and specific requirements for yield, purity, cost, and speed. Common Methods for Preparing protein sequencing Samples including:
(1) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
(2) High-performance liquid chromatography (HPLC)
(3) Capillary electrophoresis (CE)
When selecting a sample preparation method, consider factors such as experiment duration, sample purity requirements, and the quantity of protein needed. If you lack the facilities for sample preparation, MtoZ Biolabs offers comprehensive protein analysis services, including sample preparation.
2. Sample Type and Transportation
MtoZ Biolabs' comprehensive protein sequencing services can analyze samples from PVDF membranes, gel spots, gel strips, solutions, etc. For transportation, tightly seal cut protein strips and ship with ice packs. Solution samples can be vacuum-dried or freeze-dried and transported with ice packs or dry ice. Please contact us for further information.
With cutting-edge instrumentation and extensive technical expertise, MtoZ Biolabs has set the standard in the field of protein sequencing services. From basic research to industrial applications, we remain customer-focused, delivering precise and comprehensive technical support. Contact us today to embark on new discoveries in protein science!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?