Top-Down Based PTMs Characterization Service
In proteomics research and applications, understanding the complete structure and post-translational modifications (PTMs) of proteins is crucial for revealing their functions and pathological roles. Although traditional bottom-up mass spectrometry analysis is widely used for characterizing protein structures, it involves enzymatic digestion of proteins into shorter peptides, a process that often leads to the loss of critical biological information. To overcome these limitations, top-down mass spectrometry analysis has emerged, allowing scientists to conduct in-depth analysis of entire protein molecules without cutting them.
This technique not only displays the complete sequence of proteins but also accurately identifies and quantifies various post-translational modifications, such as phosphorylation and ubiquitination, while preserving the protein's original spatial structure and functional state. Moreover, top-down analysis is particularly suitable for complex biological samples, such as therapeutic monoclonal antibodies and recombinant proteins, which often contain rich forms of PTMs and sequence variants. Through comprehensive analysis of the entire protein, researchers can gain a more complete understanding of protein functions and pathological mechanisms, providing strong scientific support for the development of biomedicine.
Services at MtoZ Biolabs
MtoZ Biolabs utilizes top-down mass spectrometry to precisely identify and quantify various PTMs, such as phosphorylation and glycosylation, while revealing protein sequence integrity. Our high-end mass spectrometers, using Electrospray Ionization (ESI) or Matrix-Assisted Laser Desorption/Ionization (MALDI) techniques, effectively convert entire proteins into charged particles, which are then analyzed using advanced dissociation techniques such as Collision-Induced Dissociation (CID), Electron Capture Dissociation (ECD), or Electron Transfer Dissociation (ETD).
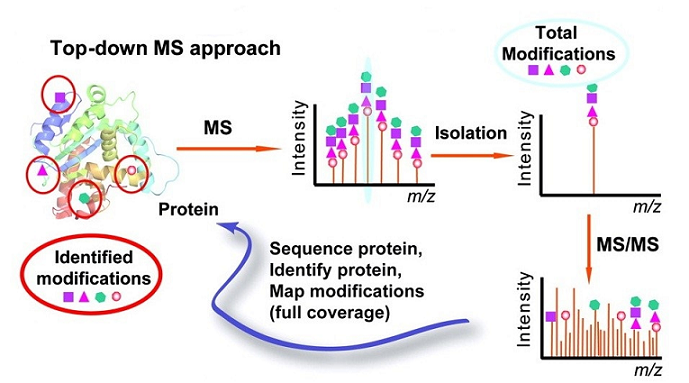
Zhang, H. et al. Circ Cardiovasc Genet. 2011.
Figure 1. Top-Down PTMs Analysis
Service Advantages
1. No Digestion Required: Our method skips the protein digestion step, preserving complete protein structures.
2. High Accuracy: Maintains all structural details and PTM data, offering high fidelity in results.
3. Sensitive Detection: Efficiently identifies crucial PTMs and protein termini.
4. Detailed PTM Analysis: Skilled at unraveling complex PTM patterns and their structural variants.
Sample Submission Requirements
1. Solid Samples: Minimum of 5 grams for plant tissue, 1 gram for animal tissue.
2. Liquid Samples: At least 1 milliliter of blood, 0.5 milliliters of serum, 10 milliliters of urine.
3. Cell and Microbial Samples: A minimum of 1×10^8 cells, or 400 milligrams dry weight for microbes.
4. Protein Samples: At least 1 milligram required.
MtoZ Biolabs' top-down PTM characterization service is designed to deliver comprehensive and precise insights, making it a leading choice for researchers and developers in the biopharmaceutical industry. For further information or to discuss specific requirements, please contact us for personalized consultation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







