De Novo Peptide Sequencing Service
De novo peptide sequencing is a method for directly deducing the amino acid sequence of peptides by analyzing mass spectrometry data, without relying on existing protein or genomic databases. De novo peptide sequencing is particularly valuable for identifying unknown proteins or novel peptides, especially when studying species or samples not represented in current databases. The de novo peptide sequencing process involves two main stages: the experimental stage and the data analysis stage. In the experimental stage, peptide mixtures are separated using high-performance liquid chromatography (HPLC), and their tandem mass spectra (MS/MS) are acquired. In the data analysis stage, the MS/MS spectra are transformed into a spectrum graph, where potential peptide sequences are scored and the most likely sequence is determined based on the highest scoring path through the graph. This process enables direct identification of peptide sequences without relying on a database.
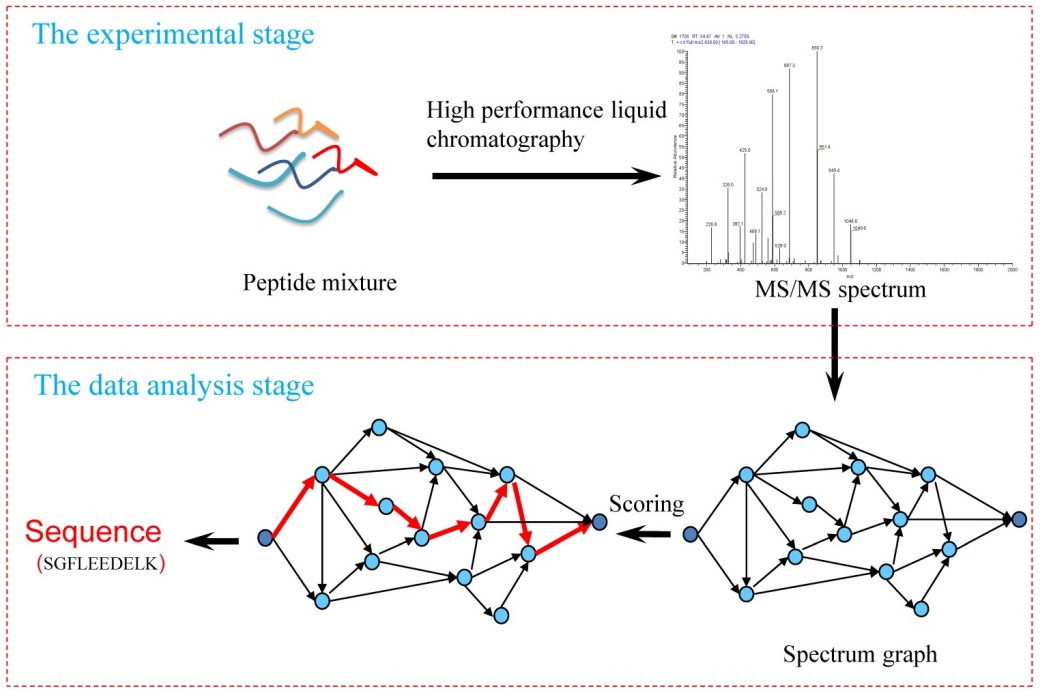
Li, C. et al. Int J Biol Sci. 2019.
Figure 1. Workflow of the De Novo Peptide Sequencing
De novo peptide sequencing addresses the limitations of traditional database-matching methods in identifying unknown sequences and is widely applied in drug development, antibody engineering, and functional research of unknown proteins. Additionally, de novo peptide sequencing provides robust support for the precise identification of low-abundance peptides and post-translational modifications, making it an essential tool for in-depth analysis of complex samples.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. With the support of high-resolution mass spectrometers and advanced liquid chromatography systems, de novo peptide sequencing service at MtoZ Biolabs ensures exceptional precision and data reliability. We utilize optimized graph-based analysis and deep learning algorithms, significantly enhancing the accuracy and efficiency of peptide sequence identification. Our technical team consists of experienced experts in mass spectrometry and proteomics, proficient in analyzing complex samples and processing data. The laboratory strictly follows optimized protocols for sample preparation and data acquisition, leveraging years of research experience to deliver high-quality and comprehensive de novo peptide sequencing solutions tailored to diverse research needs.
Service Advantages
1. Database-Free Peptide Sequence Identification
MtoZ Biolabs directly deciphers peptide sequences from mass spectrometry data without relying on any database, enabling precise identification of unknown protein sequences. This approach is particularly suited for novel samples and complex systems, providing greater flexibility and accuracy for protein sequencing.
2. Advanced Algorithms for Enhanced Accuracy
Our de novo peptide sequencing service employs advanced algorithms, including graph-based analysis, tag-based methods, and deep learning techniques, significantly improving the accuracy and efficiency of peptide sequence identification, delivering reliable protein functional data to clients.
3. High Sensitivity and Broad Applications
MtoZ Biolabs' de novo peptide sequencing service offers high sensitivity, allowing accurate identification of low-abundance peptides and modified peptides, while being compatible with various sample types. This service is ideal for protein sequencing, post-translational modification analysis, and peptide drug development, providing comprehensive peptide identification solutions for diverse research needs.
Case Study
1. Precision De Novo Peptide Sequencing Using Mirror Proteases of Ac-LysargiNase and Trypsin for Large-scale Proteomics
This study introduces a mirror protease strategy using Ac-LysargiNase and trypsin, combined with the novel algorithm pNovoM, to achieve near 100% accuracy in de novo peptide sequencing. The complementary ion series generated by these proteases enable full-length peptide sequencing with higher precision and greater coverage, outperforming existing methods in large-scale proteomics. De novo peptide sequencing service offers innovative protease strategies to achieve high accuracy and comprehensive peptide identification for diverse samples.
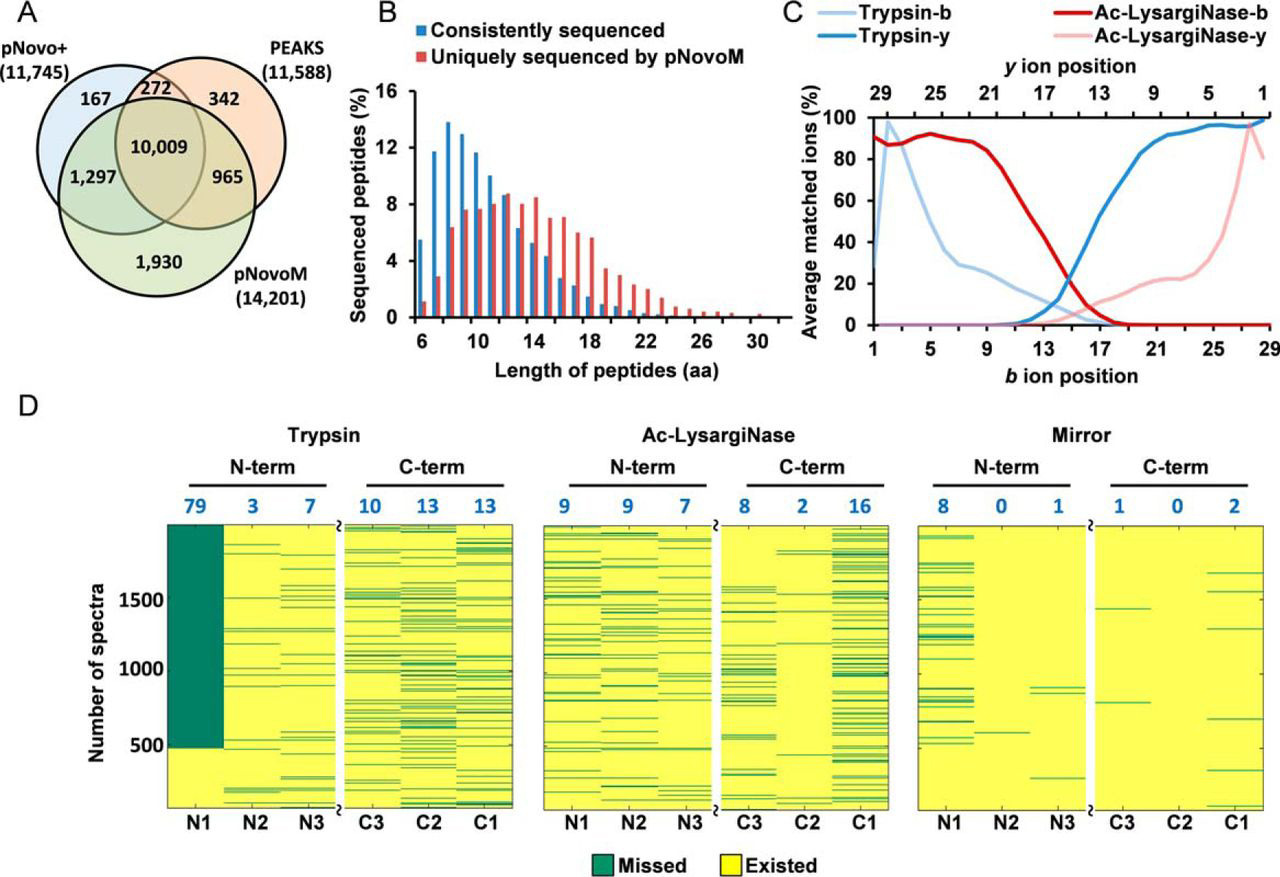
Yang, H. et al. Mol Cell Proteomics. 2019.
Figure 2. Mirror Protease Strategy for Precision De Novo Sequencing on Long Peptides
2. Accurate De Novo Peptide Sequencing Using Fully Convolutional Neural Networks
This paper introduces PepNet, a fully convolutional neural network designed for high-accuracy de novo peptide sequencing. Unlike traditional methods, PepNet significantly improves peptide-level and positional-level accuracy by analyzing MS/MS spectra, outperforming existing algorithms like PointNovo and DeepNovo. It also offers faster processing speeds and can identify spectra missed by database search engines, making it a valuable complementary tool in proteomics. De novo peptide sequencing service can utilize advanced tools to achieve high-accuracy peptide identification and accelerate large-scale proteomics analysis.
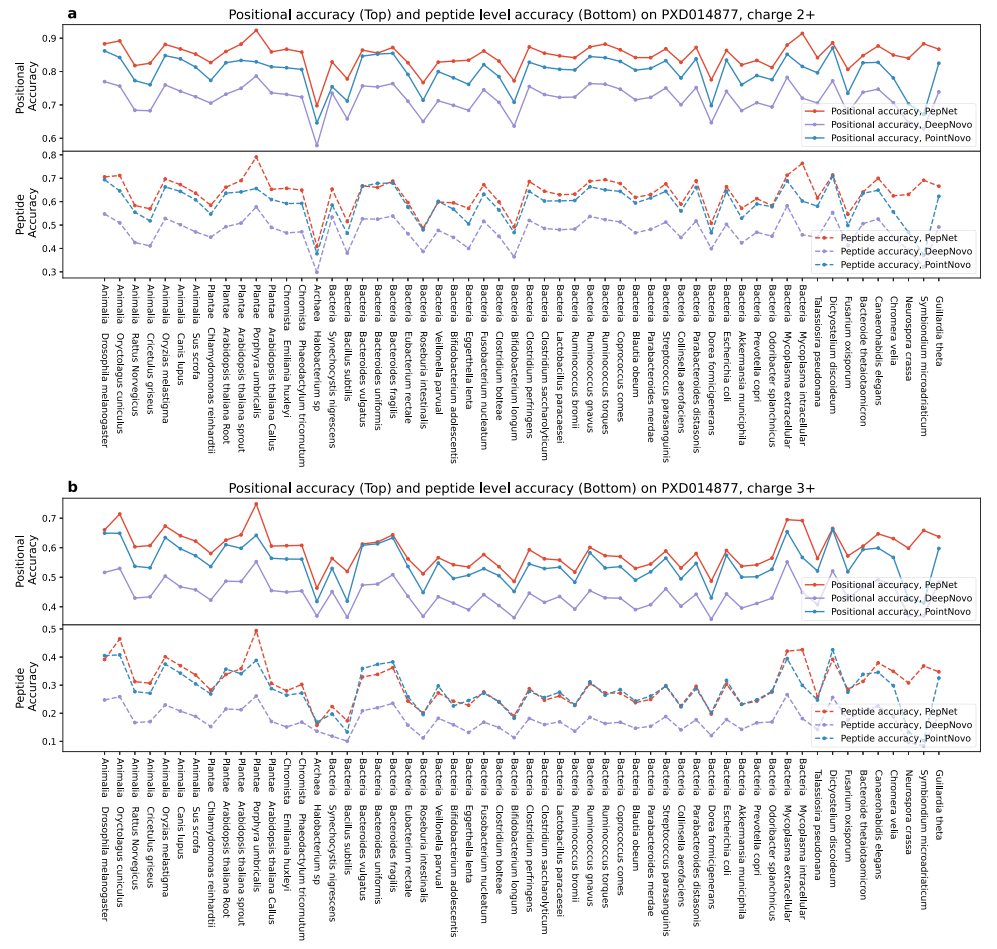
Liu, K. et al. Nat Commun. 2023.
Figure 3. Sequencing Accuracies of Peptides From Non-Human Organisms
FAQ
Q1: Does the score in de novo peptide sequencing reports refer to the confidence level of the peptide sequence? What is the typical score threshold?
Answer: The scores in de novo peptide sequencing reports typically refer to the confidence level of the peptide sequence, specifically the degree of certainty regarding the correctness of the peptide sequence. These scores are based on a comprehensive evaluation of multiple parameters, such as the match between the mass spectrometry data, ion coverage of the peptide, and signal-to-noise ratio. Higher scores generally indicate greater confidence in the results.
As for the score threshold, it may vary depending on the type of mass spectrometer, analysis software, and experimental design. The score threshold in de novo peptide sequencing service is usually determined by the laboratory based on its standards and requirements. In some cases, the threshold may be set higher to ensure accuracy; in other cases, it may be set lower to avoid missing potentially meaningful data. The choice of the threshold is typically based on prior experience, recommendations from the literature, or the default settings of the software. For instance, some software may recommend a score threshold of 1.5 or higher, but this depends entirely on the specific experimental context and objectives.
Q2: What is the difference between peptide mass spectrometry identification and de novo peptide sequencing?
Answer: The difference between peptide mass spectrometry identification and de novo peptide sequencing lies mainly in the data analysis stage.
1. Database Matching
This method identifies peptides in the sample by matching the obtained mass spectrometry data with sequences in an existing database. It relies on the availability of protein data for the corresponding species in the database.
2. De Novo Peptide Sequencing
This approach directly deduces the amino acid sequence of peptides by analyzing the fragmentation ion patterns in the mass spectrometry data, without relying on any existing database. It is suitable for cases where no relevant information is available in the database.
The sample processing steps, such as sample preparation, digestion, and ionization, are generally similar in both methods. The key difference lies in the analysis method and the data quality requirements. De novo peptide sequencing demands higher precision and complexity of the mass spectrometry data. Our de novo peptide sequencing service includes peptide mass spectrometry identification and other customized services to meet diverse research needs. Please feel free to contact us for more details.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Liquid Chromatography and Mass Spectrometry Parameters
4. The Detailed Information of De Novo Peptide Sequencing
5. Mass Spectrometry Image
6. Raw Data
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







